Annotating Gene Markers for Coding Potential and Function Predictions
In this annotated gene analysis, various annotators evaluate the coding potential and likely functions of three genes using tools like Genemark, Glimmer, Blast, and other bioinformatics resources. The annotations detail start positions, coding potentials, homologues, alignment agreements, functional regions, and other key features of the genes. Discrepancies and agreements between different annotators and tools are highlighted, providing insights into gene prediction and functional annotation processes.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
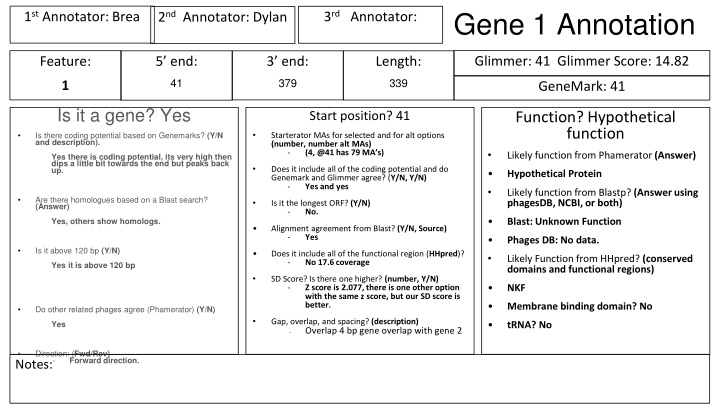
Gene 1 Annotation 1stAnnotator: Brea 3rdAnnotator: 2ndAnnotator: Dylan Glimmer: 41 Glimmer Score: 14.82 Feature: 5 end: 3 end: Length: 41 379 339 1 GeneMark: 41 Is it a gene? Yes Function? Hypothetical function Start position? 41 Is there coding potential based on Genemarks? (Y/N and description). Starterator MAs for selected and for alt options (number, number alt MAs) - (4, @41 has 79 MA s) Likely function from Phamerator (Answer) Yes there is coding potential, its very high then dips a little bit towards the end but peaks back up. Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes and yes Hypothetical Protein Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Are there homologues based on a Blast search? (Answer) Is it the longest ORF? (Y/N) - No. Yes, others show homologs. Blast: Unknown Function Alignment agreement from Blast? (Y/N, Source) - Yes Phages DB: No data. Is it above 120 bp (Y/N) Does it include all of the functional region (HHpred)? - No 17.6 coverage Likely Function from HHpred? (conserved domains and functional regions) Yes it is above 120 bp SD Score? Is there one higher? (number, Y/N) - Z score is 2.077, there is one other option with the same z score, but our SD score is better. NKF Membrane binding domain? No Do other related phages agree (Phamerator) (Y/N) Gap, overlap, and spacing? (description) Overlap 4 bp gene overlap with gene 2 Yes tRNA? No - Direction: (Fwd/Rev) - Forward direction. Notes:
Gene 2 Annotation 1stAnnotator: Omer 2ndAnnotator: Paige 3rdAnnotator: Glimmer 361, 8.46 Feature: 5 end: 3 end: Length: 2 376 1122 747 Genemarks: 376 Is it a gene? Yes Start position? 376. Function? Hypothetical Protein Starterator MAs for selected and for alt options (number, number alt MAs) - Start 2 has 77 MA Is there coding potential based on Genemarks? (Y/N and description). - Yes Likely function from Phamerator (Answer) - Unknown Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes, Yes Are there homologues based on a Blast search? (Answer) - Yes Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - Hypothetical Protein Is it the longest ORF? (Y/N) - No Alignment agreement from Blast? (Y/N, Source) - Yes, Starterator has 77 MA for bp 376 and 0 for 361 Likely Function from HHpred? (conserved domains and functional regions) - NKF Membrane binding domain? No predicted TMHs tRNA? Is it above 120 bp (Y/N) - Yes Does it include all of the functional region (HHpred)? - Yes Do other related phages agree (Phamerator) (Y/N) - Yes SD Score? Is there one higher? (number, Y/N) - -3.452 Gap, overlap, and spacing? (description) - 4 bp overlap with gene 1 and 2 bp gap with gene 3 Direction: (Fwd/Rev) - Fwd No Notes: Change called start site, based upon the MA s from starterator, still having all the coding potential, having a better SD score and Z- score. Note from Paige: GeneMark and Glimmer have different starts, so I don t think they agree. I think alignment agreement from blast should be based on NCBI or phagesdb. I am a bit confused on what you are saying in description of overlap and spacing. I am seeing a 4 overlap and 13 spacing. Everything else looks great!
Gene 3 Annotation 1stAnnotator: Dylan/Omer 2ndAnnotator: Brea 3rdAnnotator: Glimmer: 1125, 10.67 Feature: 5 end: 3 end: Length: 1461 3 1125 2585 Genemarks: 1125 Is it a gene? Yes Is there coding potential based on Genemarks? (Y/N and description). - Yes, there is good coding potential with little dips. The biggest dip starts at the very beginning but doesn t reach past the midline. There are none that go past the midline. Are there homologues based on a Blast search? (Answer) - Yes, DNAmaster shows many homologues. Although, they are all listed as hypothetical proteins Is it above 120 bp (Y/N) - Yes Do other related phages agree (Phamerator) (Y/N) - Yes Direction: (Fwd/Rev) - Fwd Start position? 1125 Function? Starterator MAs for selected and for alt options (number, number alt MAs) Likely function from Phamerator (Answer) - 114 MA s at 1125 and 4 MA s at 1131 Terminase Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - Terminase Is it the longest ORF? (Y/N) - Yes Alignment agreement from Blast? (Y/N, Source) - Yes Likely Function from HHpred? (conserved domains and functional regions) - Terminase Does it include all of the functional region (HHpred)? - Yes SD Score? Is there one higher? (number, Y/N) - Z score of 2.083, there are however they create larger gaps Gap, overlap, and spacing? (description) - 4 BP gap between gene 2 and 3 and 231 bp gap between 3 and 4 Notes:
Gene 4 Annotation 1stAnnotator: Brea 2ndAnnotator: Rylee 3rdAnnotator: Glimmer: 2815 Feature: 5 end: 3 end: Length: 4 2710 3837 1023 Genemarks: 2779 Is it a gene? Is there coding potential based on Genemarks? (Y/N and description). - Yes, there is good coding potential Are there homologues based on a Blast search? (Answer) - Yes, DNA master shows homologs listed as portal proteins Is it above 120 bp (Y/N) - Yes Do other related phages agree (Phamerator) (Y/N) - Yes Direction: (Fwd/Rev) - Fwd Start position? 2710 Function? Portal Protein Changed Start? Yes from 2815 (Both start with GTG) Likely function from Phamerator (Answer) - Head to tail connecter Starterator MAs for selected and for alt options (number, number alt MAs) - 2710, has 73 MA s - 2815 has no MA s Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - Function unknown Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - No, no Is it the longest ORF? (Y/N) - YES Likely Function from HHpred? (conserved domains and functional regions) - Portal Protein Alignment agreement from Blast? (Y/N, Source) - Yes, DNAmaster Does it include all of the functional region (HHpred)? - Yes Membrane-binding? SD Score? Is there one higher? (number, Y/N) - -6.283, No tRNA? Gap, overlap, and spacing? (description) - Makes the gap smaller Notes: Change called start site, based upon the suggested start (2815) had no MA s and not the highest shine dalgarno score. While the changed start, 2710 has 73 MA s. The new start also creates a smaller gap.
Gene 5 Annotation 1stAnnotator: Scott 2ndAnnotator: Paige 3rdAnnotator: Glimmer: 3834 (14.63) Feature: 5 end: 3 end: 5417 Length: 5 3834 1584 Genemarks: 3834 Is it a gene? Yes Is there coding potential based on Genemarks? (Y/N and description) Yes, good coding potential after 2nd start Are there homologues based on a Blast search? (Answer) Yes Is it above 120 bp (Y/N) yes Start position? 3834 Function? Major capsid protein Starterator MAs for selected and for alt options (number, number alt MAs) 78, 2 Likely function from Phamerator (Answer) Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) major capsid and protease fusion protein Y, Y Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Is it the longest ORF? (Y/N) N Major head protein Alignment agreement from Blast? (Y/N, Source) Y, Bri160 Likely Function from HHpred? (conserved domains and functional regions) Does it include all of the functional region (HHpred)? Do other related phages agree (Phamerator) (Y/N) yes Y (99.9%) Major capsid protein SD Score? Is there one higher? (number, Y/N) -1.907 (z-score: 3.088), N -Membrane binding domain? Gap, overlap, and spacing? (description) Direction: (Fwd/Rev) Fwd -tRNA? -4 bp gap, 15 bp spacing Notes: This has been reviewed by Paige.
Gene 6 Annotation 1stAnnotator: Sariah 2ndAnnotator: Scott 3rdAnnotator: Glimmer: 5421, 18.22 Feature: 5 end: 3 end: Length: 6 5421 5774 354 Genemarks: 5421, n/a Is it a gene? Yes Is there coding potential based on Genemarks? (Y/N and description). - Yes, there is good coding potential with little dips. The biggest dip starts at the very beginning and only a minor dip that goes past the midline. Are there homologs based on a Blast search? (Answer) - Yes, . Is it above 120 bp (Y/N) - Yes (354bp) Do other related phages agree (Phamerator) (Y/N) - Yes Direction: (Fwd/Rev) - Fwd Function? Head-to-Tail Adaptor Likely function from Phamerator (Answer) - Head-to-tail adaptor Start position? 5421 (ATG) Starterator MAs for selected and for alt options (number, number alt MAs) - 5421, has 80 MAs Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes, Yes Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - Head-to-tail adaptor (head-tail connector - NCBI) Is it the longest ORF? (Y/N) - Yes Alignment agreement from Blast? (Y/N, Source) - Yes, closest matches (top 10ish) have 100% alignment Likely Function from HHpred? (conserved domains and functional regions) - Hypothetical protein Does it include all of the functional region (HHpred)? - 99.14% coverage (HHpred) Adaptor protein SD Score? Is there one higher? (number, Y/N) - -4.553, No -Membrane binding domain? No Gap, overlap, and spacing? (description) - Gap: 3; Spacing: 16 -tRNA? No Notes:
Gene 7 Annotation 1stAnnotator: Dylan 2ndAnnotator: Sariah 3rdAnnotator: Glimmer: 5771 Feature: 5 end: 3 end: Length: 5771 6151 381 7 GeneMark: 5771 Is it a gene? YES Function? TBD Likely function from Phamerator (Answer) Start position? 5771 Starterator MAs for selected and for alt options (number, number alt MAs) - 80 MAs for this start. No alt MAs Is there coding potential based on Genemarks? (Y/N and description). - Yes a literal rectangle. However, it does drop off before the end of the feature/ stop codon. Tail terminator Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - All coding potential. - YES Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Minor tail protein Tail terminator protein Are there homologues based on a Blast search? (Answer) NCBI Blast via PECAAN shows numerous similar matches that have high coverage, high match, Is it the longest ORF? (Y/N) - YES Likely Function from HHpred? (conserved domains and functional regions) Alignment agreement from Blast? (Y/N, Source) - YES Is it above 120 bp (Y/N) Does it include all of the functional region (HHpred)? Phage minor tail protein YES [381] Membrane binding domain? SD Score? Is there one higher? (number, Y/N) - -5.068. 6 higher SD scores, max at -6.188 Do other related phages agree (Phamerator) (Y/N) None Gap, overlap, and spacing? (description) - Overlap of 3 at start YES Belthelas, Minima (shifted), TomBrady12 tRNA? No Direction: (Fwd/Rev) - FORWARD Notes: (I believe technically only 2 higher SD scores, highest around -4.9 and lowest of -6.188. Highest score would not include full functional region)? Most Phamerator comparisons agrees with function of Tail terminator
Gene 8 Annotation 1stAnnotator: Rylee 2ndAnnotator: Emily 3rdAnnotator: Glimmer: 6190, 13.28 Feature: 5 end: 3 end: Length: 6190 6624 435 8 Genemarks: 6190 Is it a gene? Yes Function? Major tail protein Likely function from Phamerator (Answer) Start position? 6190 Starterator MAs for selected and for alt options (number, number alt MAs) - 5, 80 MA s Is there coding potential based on Genemarks? (Y/N and description). - Yes, there is good coding potential with no real dips. There are only some small dips near the end. major tail protein, Minima Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes, Yes Are there homologues based on a Blast search? (Answer) Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Yes, there are homologues from the blast search with good E value and alignment. Is it the longest ORF? (Y/N) - Yes major tail protein from NCBI Alignment agreement from Blast? (Y/N, Source) - Yes, 100% alignment Likely Function from HHpred? (conserved domains and functional regions) Is it above 120 bp (Y/N) Yes, 435 Does it include all of the functional region (HHpred)? - YES, 99% coverage (HHpred) major tail protein Do other related phages agree (Phamerator) (Y/N) SD Score? Is there one higher? (number, Y/N) - -5.797, yes but everything else supports this start site Membrane binding domain? Yes, Minima No Direction: (Fwd/Rev) Gap, overlap, and spacing? (description) - 39 bp gap, spacing: 11 tRNA? Fwd No Notes: Secondary NFC
Gene 9 Annotation 1stAnnotator: Emily 2ndAnnotator: Brea 3rdAnnotator: Glimmer: 6637, 15.96 Feature: 5 end: 3 end: Length: 7020 9 6637 384 Genemarks: 6637, n/a Is it a gene? Yes Is there coding potential based on Genemarks? Yes, however, there is a large dip in coding potential in the middle Are there homologues based on a Blast search? Yes, PECAAN shows homologues - all listed as hypothetical proteins Is it above 120 bp (Y/N) Yes Do other related phages agree (Phamerator) (Y/N) Yes Direction: (Fwd/Rev) FWD Function? NKF Likely function from Phamerator (Answer) - NKF Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - minor tail protein phagesdB - Hypothetical protein/minor tail protein NCBI Start position? 6637 (GTG) Starterator MAs for selected and for alt options: 80 MAs for 6637, no MA for alt options Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) Yes, yes Is it the longest ORF? (Y/N) - Yes Alignment agreement from Blast? (Y/N, Source) - 100% alignment from a few results, in general - high alignments Likely Function from HHpred? (conserved domains and functional regions) Does it include all of the functional region (HHpred)? - Minor capsid protein has a 74.8031% coverage and 99.4% probability, putative tail-component has 55.9055% coverage and 99% probability) NKF SD Score? Is there one higher? (number, Y/N) - -4.294, z = 2.273, no higher Membrane binding domain? No tRNA? No Gap, overlap, and spacing? (description) Gap of 12, spacer 10 Notes: Could be a minor tail protein.
Gene 10 Annotation 1stAnnotator: Sariah 2ndAnnotator: Emily 3rdAnnotator: Glimmer: 7034, 19.32 Feature: 5 end: 3 end: Length: 7034 7354 321 10 Genemarks: 7034 Is it a gene? Yes Is there coding potential based on Genemarks? Beautiful coding potential without any dips Are there homologues based on a Blast search? (Answer) - Yes Is it above 120 bp (Y/N) - Yes, 321pb Do other related phages agree (Phamerator) (Y/N) Function? Tail Assembly Chaperone Likely function from Phamerator (Answer) Tail assembly chaperone Start position? 7034 Starterator MAs for selected and for alt options (number, number alt MAs) - Shows agreement of start site with 80 MAs Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes, Yes Is it the longest ORF? (Y/N) - Yes Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - Tail assembly chaperone (PhagesDB & NCBI) Alignment agreement from Blast? (Y/N, Source) -Majority has 100.0% alignment, a few cases of only 97+% alignment. (DNA master) Likely Function from HHpred? (conserved domains and functional regions) - Phage tail assembly chaperone protein (phage tail tube protein) -HHPRED Does it include all of the functional region (HHpred)? - Yes - Yes, compared to BoomRoasted & Anseraureola Direction: (Fwd/Rev) SD Score? Is there one higher? (number, Y/N) - SD is -3.131; Z value 2.478 Gap, overlap, and spacing? (description) - Gap of 13bp, Spacing of 10 bp Membrane bonding Site-no - FWD tRna- Nope Notes:
Gene 11 Annotation 1stAnnotator: Emily 2ndAnnotator: Sariah 3rdAnnotator: Glimmer: N/A Feature: 5 end: 3 end: Length: 7034 7470 438 11 Genemarks: N/A Is it a gene? Yes Is there coding potential based on Genemarks? Yes Are there homologues based on a Blast search? (Answer) - Yes Is it above 120 bp (Y/N) - Yes Do other related phages agree (Phamerator) (Y/N) - Yes, compared to Yubaba Direction: (Fwd/Rev) - FWD Start position? 7034 Function? Tail Assembly Chaperone Starterator MAs for selected and for alt options (number, number alt MAs) - Same start as 10 Likely function from Phamerator (Answer) Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes, N/A Tail assembly chaperone (compared to Yubaba) Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - Tail assembly chaperone phagesDB Likely Function from HHpred? (conserved domains and functional regions) - Tail assembly chaperone Is it the longest ORF? (Y/N) - Yes Alignment agreement from Blast? (Y/N, Source) - Yes Does it include all of the functional region (HHpred)? - Yes SD Score? Is there one higher? (number, Y/N) - Z score is over 2 (2.478), SD score is the closest to zero Gap, overlap, and spacing? (description) - Gap of -321, spacer 10 Notes: SD score- -3.131
Gene 12 Annotation 1stAnnotator: Dr. Grinath 2ndAnnotator: Rylee 3rdAnnotator: Glimmer: 7586 Feature: 5 end: 3 end: Length: 12 7586 9691 2106 Genemarks: agree Is it a gene? Yes Is there coding potential based on Genemarks? (Y/N and description). - Yes, there is good coding potential Are there homologues based on a Blast search? (Answer) - Yes, 25 homologues on DNAmaster with 98-100% similarity Is it above 120 bp (Y/N) - Yes Do other related phages agree (Phamerator) (Y/N) - Yes - Minima Direction: (Fwd/Rev) - Fwd Start position? 7586 Function? Tape measure protein Starterator MAs for selected and for alt options (number, number alt MAs) - Start: 4 @7586 has 80 MA's Likely function from Phamerator (Answer) - Tape measure protein Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes, Yes Is it the longest ORF? (Y/N) - Yes, and an ATG start; highest Z-score and >2 (2.782) Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - Tape measure protein (phagesDB, NCBI) Alignment agreement from Blast? (Y/N, Source) - Yes, DNAmaster, 25 hits 100% aligned Does it include all of the functional region (HHpred)? - Yes - longest ORF Likely Function from HHpred? (conserved domains and functional regions) - Tape measure protein SD Score? Is there one higher? (number, Y/N) - -2.59, No Gap, overlap, and spacing? (description) - Spacer = 7 - Gap = 115 Notes:
Gene 13 Annotation 1stAnnotator: Rylee 2ndAnnotator: Omer 3rdAnnotator: Glimmer: 9688, 12 Feature: 5 end: 3 end: Length: 9688 10647 960 13 Genemarks: 9688 Is it a gene? Yes Is there coding potential based on Genemarks? (Y/N and description). Yes, there is coding potential and it does not have any major dips just small ones. Are there homologues based on a Blast search? (Answer) Yes, has good identity and similarity both at 99%. Is it above 120 bp (Y/N) Function? minor tail protein Likely function from Phamerator (Answer) minor tail protein, BoomRoasted Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Start position? 9688 Starterator MAs for selected and for alt options (number, number alt MAs) - 1, 48 MAs Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes, Yes Is it the longest ORF? (Y/N) minor tail protein, phagesDB Yes Likely Function from HHpred? (conserved domains and functional regions) Alignment agreement from Blast? (Y/N, Source) Yes, all are 100% aligned (DNA Master) Does it include all of the functional region (HHpred)? HHpred found a structure that has to do with carbohydrates bonding. Yes, 960 Yes Do other related phages agree (Phamerator) (Y/N) Yes, BoomRoasted Direction: (Fwd/Rev) - Fwd Membrane binding domain? SD Score? Is there one higher? (number, Y/N) No -3.577, no Gap, overlap, and spacing? (description) tRNA? No Gap, 4 overlap, 12 spacing No Notes:
Gene 14 Annotation 1stAnnotator: Omer 2ndAnnotator: Rylee 3rdAnnotator: Glimmer: 10647, 9.29 Feature: 5 end: 3 end: Length: 10647 12707 2061 14 Genemarks: 10647 Is it a gene? Yes Is there coding potential based on Genemarks? (Y/N and description). Yes it has all the coding potential Are there homologues based on a Blast search? (Answer) It do be homologing Is it above 120 bp (Y/N) Yes Do other related phages agree (Phamerator) (Y/N) Yes Direction: (Fwd/Rev) - Fwd Start position? Function? Likely function from Phamerator (Minor tail protein) Starterator MAs for selected and for alt options (number, number alt MAs) - Startsite 10647 has 80 MAs Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes Minor tail protein Is it the longest ORF? (Y/N) Likely Function from HHpred? (conserved domains and functional regions) Yes Alignment agreement from Blast? (Y/N, Source) Yes Fibronectin Does it include all of the functional region (HHpred)? Yes No trna, SD Score? Is there one higher? (number, Y/N) SD score of 2.834 Gap, overlap, and spacing? (description) 0 gap with gene 13 and 3 bp spacing with gene 15 Notes:
Gene 15 Annotation 1stAnnotator: Scott 2ndAnnotator: Paige 3rdAnnotator: Glimmer: 12709 (strength: 11.63) Feature: 5 end: 3 end: Length: 12709 13287 579 15 Genemarks: 12709 Is it a gene? Yes Is there coding potential based on Genemarks? (Y/N and description). Yes Are there homologues based on a Blast search? (Answer) Yes Is it above 120 bp (Y/N) Yes Do other related phages agree (Phamerator) (Y/N) Yes Direction: (Fwd/Rev) - Fwd Start position? Function? Likely function from Phamerator (minor tail protein) Starterator MAs for selected and for alt options (number, number alt MAs) Likely function from Blastp? (Answer using phagesDB, NCBI, or both) 77 MAs (100% of EE), 23 alt (all EC) Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) minor tail protein Yes Is it the longest ORF? (Y/N) Likely Function from HHpred? (conserved domains and functional regions) Yes Alignment agreement from Blast? (Y/N, Source) Yes Does it include all of the functional region (HHpred)? Yes Short tail fiber protein No trna, SD Score? Is there one higher? (number, Y/N) -3.119; no Gap, overlap, and spacing? (description) Gap: 1; spacing: 12 Notes: Reviewed by Paige
Gene 16 Annotation 1stAnnotator: Paige 2ndAnnotator: Omer 3rdAnnotator: Glimmer: 13333, 17.58 Feature: 5 end: 3 end: Length: 13333 13596 264 16 Genemarks: 13333 Is it a gene? Start position? Function? Starterator MAs for selected and for alt options (number, number alt MAs) - 5 @ 13333 has 76 MAs Likely function from Phamerator (Answer) - None listed Is there coding potential based on Genemarks? (Y/N and description). - Yes Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes, yes Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - Hypothetical protein Is it the longest ORF? (Y/N) - Yes Are there homologues based on a Blast search? (Answer) - Yes Alignment agreement from Blast? (Y/N, Source) - Yes, with Bri160 Likely Function from HHpred? (conserved domains and functional regions) - Unknown function - Membrane binding domain? No Does it include all of the functional region (HHpred)? - Yes Is it above 120 bp (Y/N) - Yes SD Score? Is there one higher? (number, Y/N) - Found on DNA master - -5.618, 1.239 Do other related phages agree (Phamerator) (Y/N) - Yes Gap, overlap, and spacing? (description) 45 gap, 10 spacer Direction: (Fwd/Rev) - FWD - tRNA? No Notes:
Gene 17 Annotation 1stAnnotator: Omer 2ndAnnotator: Emily 3rdAnnotator: Glimmer 13620, 8.19 Feature: 5 end: 3 end: Length: 17 13620 14312 693 Genemarks: 13620 Is it a gene? Yes Start position? Function? Endolysin Starterator MAs for selected and for alt options (number, number alt MAs) - 1, 78 MA Likely function from Phamerator (Answer) - Endolysin Is there coding potential based on Genemarks? (Y/N and description). - Yes Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - Endolysin Are there homologues based on a Blast search? (Answer) - Yes Is it the longest ORF? (Y/N) - Yes Likely Function from HHpred? (conserved domains and functional regions) - Endolysin Membrane binding domain? No tRNA? No Alignment agreement from Blast? (Y/N, Source) - Yes, Starterator has 78 MA for bp 13620 Is it above 120 bp (Y/N) - Yes Does it include all of the functional region (HHpred)? - No Do other related phages agree (Phamerator) (Y/N) - Yes SD Score? Is there one higher? (number, Y/N) - -1.418, No one is higher Gap, overlap, and spacing? (description) - 288 bp gap between 16 and 17, and 32 bp overlap between 17 and 18 Direction: (Fwd/Rev) - Fwd Notes: Start position is correct based off starterator, coding potential, and Genemark/Glimmer. The most likely function of this gene is Endolysin which is what this bacteriophage uses to cleave through the cell wall of the bacteria.
Gene 18 Annotation 1stAnnotator: Brea 3rdAnnotator: 2ndAnnotator: Scott Glimmer Start: 14279 Glimmer Score: 7.46 Feature: 5 end: 3 end: Length: 14279 14545 267 18 GeneMark Start: 14312 Is it a gene? Function? Start position? Is there coding potential based on Genemarks? (Y/N and description). Starterator MAs for selected and for alt options (number, number alt MAs) - Start: 6 @14279 has 69 MA's Likely function from Phamerator (Answer) Based on the genemark hand out there is coding potential but it is spiked up and down not very constant, the coding potential for gene 18 on genemark is also combined with gene 17 for some odd reason even though there is a dip of no coding potential between the two genes. Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) No data from phamerator Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Is it the longest ORF? (Y/N) - yes Are there homologues based on a Blast search? (Answer) NCBI: Membrane Protein (100% coverage w/ e value= 3.97e^-50 Alignment agreement from Blast? (Y/N, Source) - yes Likely Function from HHpred? (conserved domains and functional regions) Does it include all of the functional region (HHpred)? Is it above 120 bp (Y/N) SD Score? Is there one higher? (number, Y/N) - 1.604, there is however it is not the longest open reading frame Yes it is over 120 bp. NKF Gap, overlap, and spacing? (description) - Gap -34 - spacing - 16 Membrane binding domain? Do other related phages agree (Phamerator) (Y/N) tRNA? Yes. Direction: (Fwd/Rev) - Notes: I think 14309 might be a better start. Pros: better SD score, better gap, spacing and alignment are the same, still contains most of the coding potential. Cons: less common MA, not LORF, doesn t match Glimmer or GeneMark (though they don t agree either) Talked over with Dr. Thomas better start site 14279. Forward
Gene 19 Annotation 1stAnnotator: Scott 2ndAnnotator: Brea 3rdAnnotator: Glimmer: 14542 Feature: 19 5 end: 3 end: 14766 Length: 225 14542 Genemarks: 14542 Is it a gene? Yes Start position? Function? Unknown function Likely function from Phamerator (Answer) - Unknown function Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Starterator MAs for selected and for alt options (number, number alt MAs) Is there coding potential based on Genemarks? (Y/N and description). - Yes 69 MA, 11 alt Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes Are there homologues based on a Blast search? (Answer) - Yes Is it the longest ORF? (Y/N) - Yes Holin OR membrane protein Likely Function from HHpred? (conserved domains and functional regions) - Holin Membrane binding domain? one tRNA? no Alignment agreement from Blast? (Y/N, Source) - Yes Is it above 120 bp (Y/N) - Yes Does it include all of the functional region (HHpred)? - Yes Do other related phages agree (Phamerator) (Y/N) - Yes SD Score? Is there one higher? (number, Y/N) - -2.414 Gap: -4bp, overlap, and spacing: 11 Direction: (Fwd/Rev) - Fwd Notes: has good matches on BlastP for membrane protein and holin, but does not meet the requirements listed on official function list. Most holins have multiple transmembrane regions, but this gene only has one.
1stAnnotator: Sariah 2ndAnnotator: Paige Gene 20 Annotation 3rdAnnotator: Glimmer 15048 9.28 Feature: 5 end: 3 end: Length: 14836 15048 213 20 Genemarks: 15048 Is it a gene? Is there coding potential based on Genemarks? Very good coding potential with no dips from start to stop Are there homologues based on a Blast search? Yes Is it above 120 bp (Y/N) Yes (213bp) Do other related phages agree (Phamerator) (Y/N) Yes, Minima & Sara Direction: (Fwd/Rev) Reverse Function? Start position? 15048 (ATG) Starterator MAs for selected and for alt options: Starterator for Feature 21 (rather than 20) shows agreement with 78 MAs Likely function from Phamerator (Answer) - Lsr2-like dna binding protein (compared with BoomRoasted) Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) Yes, Yes Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - Lsr2-like dna binding protein (PhagesDB, NCBI in agreement) Is it the longest ORF? (Y/N) - Yes Alignment agreement from Blast? (Y/N, Source) - Majority has 100.0% alignment, cases of only 78- 92% alignment. (DNA master) - Ranges from 71-98% (PECAAN- NCBI Blast) Likely Function from HHpred? (conserved domains and functional regions) Protein lsr2; anti-parallel beta sheet, dimer, DNA Binding Protein (Mycobacterium tuberculosis) Does it include all of the functional region (HHpred)? - Yes SD Score? Is there one higher? (number, Y/N) - Z score: 2.681; No higher - SD score: -3.627 Gap, overlap, and spacing? (description) Membrane binding domain? no tRNA? no Gap:2; Spacing 12 Notes from Paige: I got SD score -2.791. Z-value 2.648. Everything else looked good.
Gene 21 Annotation 1stAnnotator: Dylan 2ndAnnotator: Sariah/Rylee 3rdAnnotator: Glimmer: 15,494 Feature: 5 end: 3 end: Length: 15,557 15,501 507bp 21 GeneMark: 15,557 Is it a gene? Is there coding potential based on Genemarks? (Y/N and description). Decent coding potential. One minor dips below 0.5. Are there homologues based on a Blast search? (Answer) Yes Is it above 120 bp (Y/N) Yes, 507bp Do other related phages agree (Phamerator) (Y/N) Yes, compared to Yubaba and BoomRoasted Direction: (Fwd/Rev) Reverse Function? Helix-turn-helix binding domain Start position? 15,557 Starterator MAs for selected and for alt options (number, number alt MAs) - 73 MAs at 15,557 - 1 MA @ 15,551 - 12 MAs @ 15,494 Likely function from Phamerator (Answer) Helix-turn-helix binding protein - Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Helix-turn-helix binding domain (PhagesDB, HHPred & NCBI Blast) Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes? (hard to tell through Genemark), no, no - Is it the longest ORF? (Y/N) - No Likely Function from HHpred? (conserved domains and functional regions) Helix-turn-helix binding domian Alignment agreement from Blast? (Y/N, Source) - Has okay alignment all over 80% (btwn 85-97) Does it include all of the functional region (HHpred)? - yes Membrane binding domain? SD Score? Is there one higher? (number, Y/N) - SD Score: -7.110 - Z score: 0.505 no tRNA? No Gap, overlap, and spacing? (description) - Gap: 81; Spacing: 12 Notes: For the start site we would not use 15,494 because not only does it have less MAs but also because the gap is almost twice as large (144 bp) as the gap between genes when using 15,557 as the starting site.
Gene 22 Annotation 1stAnnotator: Rylee 2ndAnnotator: 3rdAnnotator: Glimmer: 15869, 4.91 Feature: 5 end: 3 end: Length: 22 15869 15639 231 Genemarks: 15869 Is it a gene? Yes Is there coding potential based on Genemarks? (Y/N and description). - Yes, but there is a large dip in the middle of it. Are there homologues based on a Blast search? (Answer) - Yes, good E value and alignment. Is it above 120 bp (Y/N) - Yes, 231 Do other related phages agree (Phamerator) (Y/N) Yes, TeddyBoy and BoomRoasted Direction: (Fwd/Rev) - REV Function?helix-turn-helix DNA binding domain protein Start position? 15869 Starterator MAs for selected and for alt options (number, number alt MAs) Likely function from Phamerator (Answer) 5, 80 MA s helix-turn-helix DNA binding domain protein Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - YES, Yes Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - helix-turn-helix DNA binding domain protein (PhagesDB) Is it the longest ORF? (Y/N) - Yes Alignment agreement from Blast? (Y/N, Source) - Yes, 100% alignment Likely Function from HHpred? - helix-turn-helix DNA binding domain protein Does it include all of the functional region (HHpred)? - Yes, all around 90% coverage Membrane binding domain? SD Score? Is there one higher? (number, Y/N) - -6.237, No No Gap, overlap, and spacing? (description) tRNA? 145 gap, 15 spacing, no overlap No Notes:
Gene 23 Annotation 1stAnnotator: Emily 2ndAnnotator: Omer 3rdAnnotator: Glimmer: 16378, 4.7 Feature: 5 end: 3 end: Length: 16378 16764 387 23 Genemarks: 16582 Is it a gene? Is there coding potential based on Genemarks? Yes, start position begins before CP, coding potential starts very hesitant Are there homologues based on a Blast search? Yes Is it above 120 bp (Y/N) yes Do other related phages agree (Phamerator) (Y/N) Some agree (Yubaba) some disagree (Minima) Direction: (Fwd/Rev) FWD Function? Hypothetical protein Likely function from Phamerator (Answer) - NKF Start position? 16378 Starterator MAs for selected and for alt options: Start @16378 has 58 MA's, no other starts with MA Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) Yes, no Likely function from Blastp? (Answer using phagesDB, NCBI, or both) - NCBI hypothetical function - phagesDB NKF Is it the longest ORF? (Y/N) - Yes Alignment agreement from Blast? (Y/N, Source) - Yes, two with 100, one with 99+ Likely Function from HHpred? (conserved domains and functional regions) Does it include all of the functional region (HHpred)? - No SD Score? Is there one higher? (number, Y/N) - Z = 1.983, no higher -5.399 NKF Gap, overlap, and spacing? (description) Membrane binding domain? No tRNA? No Gap = 508 Spacer = 8 Notes:
Gene 24 Annotation 1stAnnotator: Paige 2ndAnnotator: Sariah All checks out other than gap anomaly 3rdAnnotator: Glimmer: 16854, 11.03 Feature: 5 end: 3 end: Length: 24 16854 17072 219 Genemarks: 16863 Is it a gene? Is there coding potential based on Genemarks? (Y/N and description). - Yes Are there homologues based on a Blast search? (Answer) - Yes Is it above 120 bp (Y/N) - Yes Do other related phages agree (Phamerator) (Y/N) Yes Direction: (Fwd/Rev) - FWD Function? Start position? Starterator MAs for selected and for alt options (number, number alt MAs) Likely function from Phamerator (Answer) - Helix-turn-helix DNA binding domain protein 11 @16854 has 3 MAs Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Yes, no Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Helix-turn-helix DNA binding domain protein Is it the longest ORF? (Y/N) - Yes - Alignment agreement from Blast? (Y/N, Source) - Yes Likely Function from HHpred? (conserved domains and functional regions) - Helix-turn-helix DNA binding domain protein Does it include all of the functional region (HHpred)? - Yes? SD Score? Is there one higher? (number, Y/N) - -6.171 z- score 0.964, yes - Membrane binding domain? Gap, overlap, and spacing? (description) 89 gap is higher than it should be, 13 spacer - tRNA? No Notes: Gap is larger than 50, which could be a problem. I reviewed it and I think it is fine, but it may need further review to make sure.
Gene 25 Annotation 1stAnnotator: Paige 3rdAnnotator: 2ndAnnotator: Scott Glimmer: 17069, 5.76 Feature: 5 end: 3 end: Length: 17069 17368 300 25 GeneMark: 17069 Is it a gene? Is there coding potential based on Genemarks? (Y/N and description). - Y Are there homologues based on a Blast search? (Answer) - Y Is it above 120 bp (Y/N) Y Do other related phages agree (Phamerator) (Y/N) Y Direction: (Fwd/Rev) - FWD Start position? Function? Starterator MAs for selected and for alt options (number, number alt MAs) - 21 @ 17069 has 81 MAs Likely function from Phamerator (Answer) Does it include all of the coding potential and do Genemark and Glimmer agree? (Y/N, Y/N) - Y?, Y HNH endonuclease Likely function from Blastp? (Answer using phagesDB, NCBI, or both) Is it the longest ORF? (Y/N) - N HNH endonuclease Alignment agreement from Blast? (Y/N, Source) - Y, BoomRoasted Likely Function from HHpred? (conserved domains and functional regions) Does it include all of the functional region (HHpred)? - Y? High E-value, unreliable SD Score? Is there one higher? (number, Y/N) - -4.502, 1.796, Y Membrane binding domain? no Gap, overlap, and spacing? (description) - Overlap 4, 15 spacer tRNA? No Notes: DNA repair