Bioengineering Design for Cyanide Degradation by Alexander Popescu and German Homeschool Team
"Discover a bioengineering solution to degrade cyanide efficiently and safely in contaminated groundwater using genes from Pseudomonas pseudoalcaligenes, addressing environmental concerns. Explore the system and parts design for effective cyanide biodegradation."
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
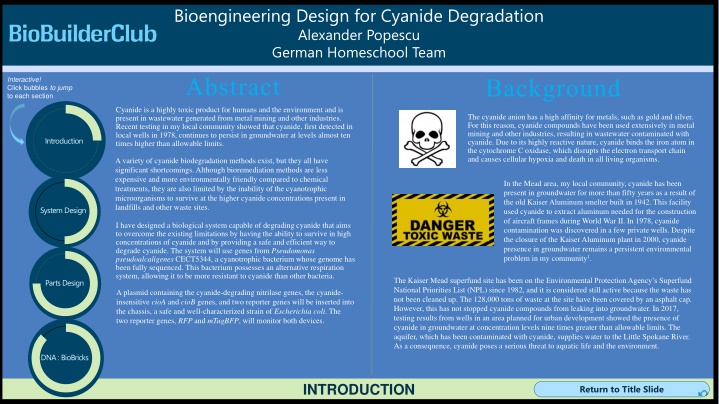
Bioengineering Design for Cyanide Degradation Alexander Popescu German Homeschool Team Bioengineering Design for Cyanide Degradation Alexander Popescu German Homeschool Team Abstract Background Interactive! Click bubbles to jump to each section Cyanide is a highly toxic product for humans and the environment and is present in wastewater generated from metal mining and other industries. Recent testing in my local community showed that cyanide, first detected in local wells in 1978, continues to persist in groundwater at levels almost ten times higher than allowable limits. The cyanide anion has a high affinity for metals, such as gold and silver. For this reason, cyanide compounds have been used extensively in metal mining and other industries, resulting in wastewater contaminated with cyanide. Due to its highly reactive nature, cyanide binds the iron atom in the cytochrome C oxidase, which disrupts the electron transport chain and causes cellular hypoxia and death in all living organisms. Introduction A variety of cyanide biodegradation methods exist, but they all have significant shortcomings. Although bioremediation methods are less expensive and more environmentally friendly compared to chemical treatments, they are also limited by the inability of the cyanotrophic microorganisms to survive at the higher cyanide concentrations present in landfills and other waste sites. In the Mead area, my local community, cyanide has been present in groundwater for more than fifty years as a result of the old Kaiser Aluminum smelter built in 1942. This facility used cyanide to extract aluminum needed for the construction of aircraft frames during World War II. In 1978, cyanide contamination was discovered in a few private wells. Despite the closure of the Kaiser Aluminum plant in 2000, cyanide presence in groundwater remains a persistent environmental problem in my community1. System Design I have designed a biological system capable of degrading cyanide that aims to overcome the existing limitations by having the ability to survive in high concentrations of cyanide and by providing a safe and efficient way to degrade cyanide. The system will use genes from Pseudomonas pseudoalcaligenes CECT5344, a cyanotrophic bacterium whose genome has been fully sequenced. This bacterium possesses an alternative respiration system, allowing it to be more resistant to cyanide than other bacteria. The Kaiser Mead superfund site has been on the Environmental Protection Agency s Superfund National Priorities List (NPL) since 1982, and it is considered still active because the waste has not been cleaned up. The 128,000 tons of waste at the site have been covered by an asphalt cap. However, this has not stopped cyanide compounds from leaking into groundwater. In 2017, testing results from wells in an area planned for urban development showed the presence of cyanide in groundwater at concentration levels nine times greater than allowable limits. The aquifer, which has been contaminated with cyanide, supplies water to the Little Spokane River. As a consequence, cyanide poses a serious threat to aquatic life and the environment. Parts Design A plasmid containing the cyanide-degrading nitrilase genes, the cyanide- insensitive cioA and cioB genes, and two reporter genes will be inserted into the chassis, a safe and well-characterized strain of Escherichia coli. The two reporter genes, RFP and mTagBFP, will monitor both devices. DNA : BioBricks INTRODUCTION INTRODUCTION Return to Title Slide Return to Title Slide
Bioengineering Design for Cyanide Degradation Alexander Popescu German Homeschool Team Bioengineering Design for Cyanide Degradation Alexander Popescu German Homeschool Team Interactive! Click bubbles to jump to each section System Level Design Device Level Design The system uses a safe and well characterized strain of E. coli as a chassis. The design includes two separate devices in order to ensure optimal efficiency and monitoring of the system. The system needs to accomplish the following goals: 1. Efficiently biodegrade cyanide 2. Have the capability to resist higher concentrations of cyanide 3. Be safe for humans and the environment 4. Be easy to monitor The first device has the role of ensuring the survival of the transformed E. coli. This device contains the cioA and cioB genes, which are clustered together and encode a quinol oxidase insensitive to cyanide5,6. Introduction An essential component of our system is the cyanide degradation genes and the cyanide insensitive genes from P. pseudoalcaligenes. I decided to use genes from P. pseudoalcaligenes, a bacterium capable of degrading cyanide, for the following reasons: 1. P. pseudoalcaligenes utilizes cyanide as its only nitrogen source and was able to tolerate up to thirty mM free cyanide2. 2. The complete genome of P. pseudoalcaligenes was sequenced in 20143. 3. This bacterium possesses a cyanide-insensitive respiration system cytochrome, making it more resistant to cyanide than other bacteria. DEVICE #1 CYANIDE System Design CN Resistance Red Fluorescence The second device degrades cyanide and contains the nit1C gene, which encodes the nitrilase enzyme, and the mqo gene, which encodes malate:quinone oxidoreductase, the enzyme important for the reaction of cyanide and oxaloacetate to from a nitrile in the first step of the nitrilase pathway. My system incorporates the nitrilase nit1C gene cluster, which is necessary for the degradation of cyanide and aliphatic nitrile4. The cioA and cioB genes encode a quinol oxidase insensitive to cyanide and ensure the survival of the system in the presence of high concentrations of cyanide5,6. Parts Design The system contains two reporter genes, the RFP gene and the mTagBFP gene. The purpose of these genes is to monitor the system in order to signal if it is functioning properly. DEVICE #2 CYANIDE CN Biodegradation Blue DNA : BioBricks Fluorescence SYSTEM LEVEL DESIGN Return to Title Slide SYSTEM LEVEL DESIGN Return to Title Slide
Bioengineering Design for Cyanide Degradation Alexander Popescu German Homeschool Team Bioengineering Design for Cyanide Degradation Alexander Popescu German Homeschool Team Interactive! Click bubbles to jump to each section Parts Level Design Both devices contain a BioBrick that includes a T7 constitutive promoter, an RBS, and a transcriptional regulator. DNA BioBrick Parts Design Diagram Number Part The T7 promoter leads to the overexpression of important enzymes necessary for the system, contributing to the fast and efficient response of this assembly when cyanide is present in the environment. The T7 promoter binds RNA polymerase, promoting transcription of the mRNA, a process which can be enhanced or inhibited by the transcriptional regulator. T7 constitutive promoter, RBS, and a transcriptional regulator (Part:BBa_K944012) Introduction 1 ORF for cioA gene (Part:BBa_K944003) 2 ORF for cioB gene (Part:BBa_K944004) 3 System Design ORF for RFP gene (Part:BBa_K1399003) 4 5 Forward terminator (Part:BBa_B0010) T7 constitutive promoter, RBS, and a transcriptional regulator (Part:BBa_K944012) 6 Cyanide degradation genes (nit1C gene cluster; GenBank: JF748722.1, https://www.ncbi.nlm.nih.gov/nuccore/JF748722.1) Parts Design 7 mqo gene for malate-quinone oxidoreductase (Part:BBa_M1102) 8 ORF for mTagBFP gene (Part:BBa_K592100) 9 DNA : BioBricks 10 Forward terminator (Part:BBa_B0010) PARTS LEVEL DESIGN Return to Title Slide PARTS LEVEL DESIGN Return to Title Slide
Bioengineering Design for Cyanide Degradation Alexander Popescu German Homeschool Team Bioengineering Design for Cyanide Degradation Alexander Popescu German Homeschool Team Interactive! Click bubbles to jump to each section Acknowledgements 8. Luque-Almagro, V. M., Huertas, M.-J., Martinez-Luque, M., Moreno-Vivian, C., Roldan, M. D., Garcia-Gil, L. J., Blasco, R. (2005). Bacterial Degradation of Cyanide and Its Metal Complexes under Alkaline Conditions. Applied and Environmental Microbiology, 71(2), 940 947. doi: 10.1128/aem.71.2.940-947.2005 9. Sung, Y. C., & Fuchs, J. A. (1992). The Escherichia coli K-12 cyn operon is positively regulated by a member of the lysR family. Journal of Bacteriology, 174(11), 3645 3650. doi: 10.1128/jb.174.11.3645-3650.1992 10.Luque-Almagro, V. M., Huertas, M.-J., Saez, L. P., Luque-Romero, M. M., Moreno-Vivian, C., Castillo, F., Blasco, R. (2008). Characterization of the Pseudomonas pseudoalcaligenes CECT5344 Cyanase, an Enzyme That Is Not Essential for Cyanide Assimilation. Applied and Environmental Microbiology, 74(20), 6280 6288. doi: 10.1128/aem.00916-08 11.IGEM Team Panama: Genetically Modified E coli as an Alternative Biosensor for Cyanide and Cyanide Compounds [Internet]. 2012 November. Available from: http://2012.igem.org/PROJECT.html 12.Wibberg, D., Luque-Almagro, V. M., Ige o, M. I., Bremges, A., Rold n, M. D., Merch n, F., Schl ter, A. (2014). Complete genome sequence of the cyanide-degrading bacterium Pseudomonas pseudoalcaligenes CECT5344. Journal of Biotechnology, 175, 67 68. doi: 10.1016/j.jbiotec.2014.02.004 13.Luque-Almagro, V. M., Cabello, P., S ez, L. P., Olaya-Abril, A., Moreno-Vivi n, C., & Rold n, M. D. (2017). Exploring anaerobic environments for cyanide and cyano-derivatives microbial degradation. Applied Microbiology and Biotechnology, 102(3), 1067 1074. doi: 10.1007/s00253- 017-8678-6 14.Kuldell N, Bernstein Ingram K, Hart K.(2015) Biobuilder. Sebastopol, CA: OReilly 15. Sharma, M., Akhter, Y. & Chatterjee, S. (2019). A review on remediation of cyanide containing industrial wastes using biological systems with special reference to enzymatic degradation. World J Microbiol Biotech 35, 70 (2019) doi:10.1007/s11274-019-2643-8 16.Phillips, T. (2019, November 20). Top 6 Reasons E. coli Is Used for Gene Cloning. Retrieved from https://www.thoughtco.com/top-reasons-e-coli-is-used-for-gene-cloning-375742 17. Sowa,T. (Oct 2013). The next phase: Kaiser s Mead smelting plant undergoes demolition The Spokesman in Review. 18.Figueira, Marianne & Ciminelli, V. & Andrade, Mabel & Linardi, Valter. (1996). Cyanide degradation by an Escherichia coli strain. Canadian journal of microbiology. 42. 519-23. 10.1139/m96-070. I am grateful to Dr. Patricia Silveyra for her mentorship and for the support and feedback she has provided for this project. I would also like to thank the BioBuilder team for all their help and suggestions. Introduction References 1. Hill, K. New cyanide cleanup plan proposed for former Kaiser Aluminum smelter site in Mead The Spokesman in Review, April 2019. 2. Karamba et al. Biological remediation of cyanide: A review. Biotropia 2015. Vol 22. No2, 2015: 151-163 DOI:10.11598/btb.2015.22.2.393 3. Luque-Almagro, V. M., Moreno-Vivi n, C., & Rold n, M. D. (2016). Biodegradation of cyanide wastes from mining and jewellery industries. Current Opinion in Biotechnology, 38, 9 13. doi: 10.1016/j.copbio.2015.12.004 4. Estepa, J.Luque-Almagro, V. Cobos, I. Escribano, M. Mart nez-Luque, M. Moreno-Vivi n, C. Rold n, M. (2012). The nit1C gene cluster of Pseudomonas pseudoalcaligenes CECT5344 involved in assimilation of nitriles is essential for growth on cyanide. Environmental microbiology reports. 4. 326-34. doi:10.1111/j.1758-2229.2012.00337.x. 5. Quesada, A., Guijo, M. I., Merchan, F., Blazquez, B., Igeno, M. I., & Blasco, R. (2007). Essential Role of Cytochrome bd-Related Oxidase in Cyanide Resistance of Pseudomonas pseudoalcaligenes CECT5344. Applied and Environmental Microbiology, 73(16), 5118 5124. doi: 10.1128/aem.00503-07 6. S ez, L. P., Cabello, P., Ib ez, M. I., Luque-Almagro, V. M., Rold n, M. D., & Moreno- Vivi n, C. (2019). Cyanate Assimilation by the Alkaliphilic Cyanide-Degrading Bacterium Pseudomonas pseudoalcaligenes CECT5344: Mutational Analysis of the cyn Gene Cluster. International Journal of Molecular Sciences, 20(12), 3008. doi: 10.3390/ijms201230 7. Cabello, P., Luque-Almagro, V. M., Olaya-Abril, A., S ez, L. P., Moreno-Vivi n, C., & Rold n, M. D. (2018). Assimilation of cyanide and cyano-derivatives by Pseudomonas pseudoalcaligenes CECT5344: from omic approaches to biotechnological applications. FEMS Microbiology Letters, 365(6). doi: 10.1093/femsle/fny032 System Design Parts Design DNA : BioBricks DNA BioBrick PARTS DNA BioBrick PARTS Return to Title Slide Return to Title Slide