Blunt and Cohesive End Cloning Techniques
Blunt and cohesive end cloning are essential DNA manipulation methods using enzymes like Klenow Fragment and T4 DNA Polymerase. Blunting involves preparing DNA fragments for ligation by removing unpaired bases, while cohesive end cloning helps in creating compatible ends for DNA manipulation and research purposes. Learn about the applications and benefits of these techniques in molecular biology experiments.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
Klenow Enzyme DNA Polymerase I, Klenow Fragment (Large Fragment) is a proteolytic product of E. coli DNA Polymerase I, which possesses the 5' 3' polymerase and 3' 5' exonuclease activities of intact DNA Polymerase I, but lacks the 5' 3' exonuclease activity of full-length DNA Polymerase I. Because the 5' 3' exonuclease activity of DNA polymerase I from E. coli makes it unsuitable for many applications, the Klenow fragment, which lacks this activity, can be very useful in research. The Klenow fragment is extremely useful for research- based tasks such as: 1. Synthesis of double-stranded DNA from single-stranded templates 2. Filling in receded 3' ends of DNA fragments to make 5' overhang blunt ..To make blunt 3. Digesting away protruding 3' overhangs .To make blunt 4. Preparation of radioactive DNA probes
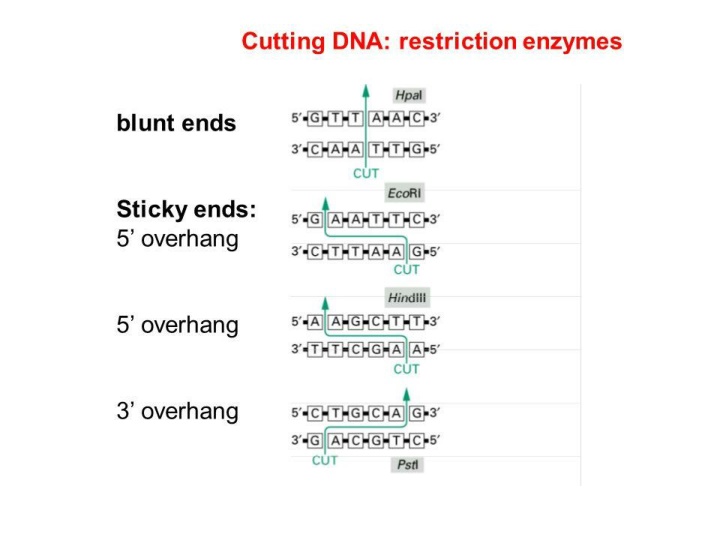
Blunting a fragment of DNA involves the removal or fill-in of any unpaired, overhanging bases. This process is often used to prepare fragments for ligation with other blunt-ended DNA, or fragments with non-compatible ends Fill in 5 -3 Fill in 5 -3 Degrades 3 -5
The Klenow fragment was also the original enzyme used for greatly amplifying segments of DNA in the polymerase chain reaction (PCR) process,before being replaced by thermostable enzymes such as Taq polymerase. exo- Klenow fragment (Lacking both exonuclease activity)
T4 DNA Polymerase T4 DNA Polymerase catalyzes the synthesis of DNA in the 5 3 direction and requires the presence of template and primer. This enzyme has a 3 5 exonuclease activity which is much more active than that found in DNA Polymerase I (E. coli). Unlike E. coli DNA Polymerase I, T4 DNA Polymerase does not have a 5 3 exonuclease function. Applications: 1. 3 overhang removal to form blunt ends. 2. 5 overhang fill-in to form blunt ends. 3. Single strand deletion subcloning. 4. Second strand synthesis in site-directed mutagenesis. 5. Probe labeling using replacement synthesis.
Cloning vs Subcloning Cloning refers to the process of creating clones of organisms or copies of cells or DNA fragments while subcloning refers to a technique used to move a particular DNA sequence from a parent vector to a destination vector. Thus, this is the main difference between cloning and subcloning.
Arthur Kornberg (March 3, 1918 October 26, 2007) was an American biochemist who won the Nobel Prize in Physiology or Medicine 1959 for his discovery of "the mechanisms in the biological synthesis of deoxyribonucleic acid (DNA)" together with Spanish biochemist and physician Severo Ochoa of New York University. (DNA Polymerase I) Thomas American biochemist who was the first person to purify and characterise DNA polymerase II and DNA polymerase III. He is currently a professor of biochemistry and biophysics at the University of California, San Francisco, and is working melanogaster development. Bill Kornberg is an on Drosophila
Roger Kornberg (born April 24,1947) is an American biochemist and professor of structural biology at Stanford University School of Medicine. Kornberg was awarded the Nobel Prize in Chemistry in 2006 for his studies of the process by which genetic information from DNA is copied to RNA, "the molecular basis of eukaryotic transcription.
DNA Modifying Enzymes 1. Composition Modifiers Nucleases Methylases and Demethylases Phosphatases and Kinases Polymerases Ligases 2. Topology Modifiers Topoisomerases
Nucleases: A nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Exonucleases digest nucleic acids from the ends. Endonucleases act on regions in the middle of target molecules. They are further subcategorized as deoxyribonucleases and ribonucleases. The former acts on DNA, the latter on RNA. Nuclease activities are integral parts of DNA replication; the 5 to 3 exo- and endonucleases are needed to remove RNA primers and the 3 to 5 exonuclease for proofreading. Two other major DNA metabolic processes, recombination and repair, are initiated by nucleases. Nuclease activity is also required for structural alterations of nucleic acids, for example, topoisomerization site-specific recombination and RNA splicing during which a phosphodiester bond is temporarily broken and reformed after strand passing or transfer to a new target.
In addition, nuclease activities are essential in RNA processing, maturation, and RNA interference. RNA and DNA degradation is an essential component of microbial defense mechanisms. Nucleases are even essential for programmed cell death . Defective DNase and RNase activities have been associated with various autoimmune diseases due to incomplete removal of endogenously produced nucleic acids A nuclease is a phosphodiesterase that cleaves one of the two bridging P-O bonds, 3 or 5 , in a nucleic acid polymer
Exonucleases, which remove one nucleotide at a time from the end of a strand, can be further divided to two groups by the 5 to 3 versus 3 to 5 polarity.
Exonuclease usually attack either the 5 or 3 ends. 1. Source: Escherichia coli Exonuclease I breaks apart single-stranded DNA in a 3' 5' direction, releasing deoxyribonucleoside 5'-monophosphates one after another. ExoI ssDNA 2. Source: Escherichia coli Exonuclease III breaks apart double-stranded DNA in a 3' 5' direction, releasing deoxyribonucleoside 5'-monophosphates one after another. ExoIII dsDNA
3. WRN Exonuclease Werner Syndrome>>>>>>autosomal recessive disorder 3' 5' direction dsDNA 4. TREX1 and TREX2 are the major mammalian 3 5 exonucleases and also prefer single stranded DNA substrate .. ssDNA Aicardi-Gouti res syndrome, a disorder that involves severe brain dysfunction (encephalopathy) >>>>>>>>>>>autosomal recessive disorder 5. RNaseT 3' 5' direction ssRNA 6. 5 -3 exoribonuclease 1 (Xrn1) is a protein that in humans is encoded by the XRN1 gene. Xrn1 hydrolyses RNA in the 5 to 3 direction.
>>>>>>>>>>>>>Nuclease BAL31 exonuclease degrades both 3' and 5' termini of duplex DNA without generating internal scissions. BAL 31 Nuclease acts as an exonuclease, degrading double-stranded DNA and RNA from both 5'-phosphate and 3'-hydroxyl termini. This enzyme also possesses single- stranded DNA and RNA endonuclease activity and is capable of cleaving at DNA/RNA nicks and gaps. It requires Ca2+as an essential cofactor for its exonuclease and endonuclease activities. Source: Alteromonas espejiana
Members of the Flap endonuclease 1 (FEN1) family have the 5 to 3 exonuclease activity in addition to the endonuclease activity. Mre11, which is involved in DNA double-strand break (DSB) repair, has both endo and 3 to 5 exo-nuclease activities Q1. Can exonuclease cut plasmid?