Comparative Analysis of Evolutionary Parameters in Giraffe and Okapi Vision Genes
This presentation compares three evolutionary parameters (dN, dS, dN/dS) between giraffe and okapi in a set of vision genes using the free-ratio model of the PAML program. The images illustrate the differences in nonsynonymous and synonymous substitutions, as well as the ratio of nonsynonymous to synonymous substitutions, shedding light on the evolutionary patterns in these two related species.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
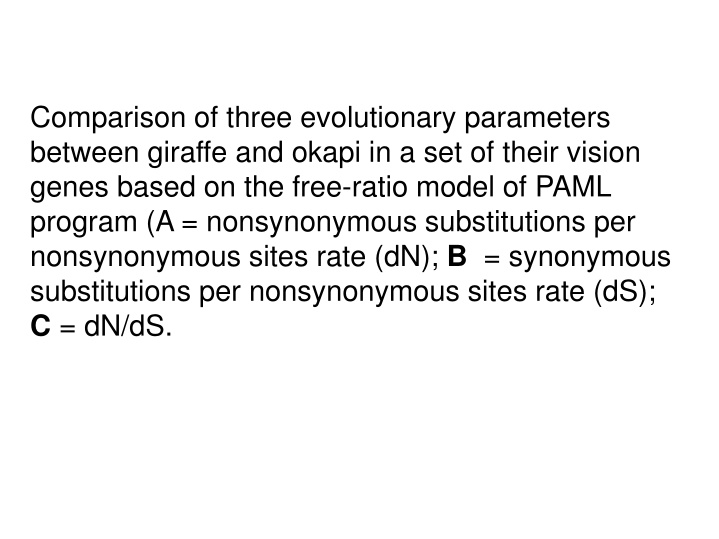
Comparison of three evolutionary parameters between giraffe and okapi in a set of their vision genes based on the free-ratio model of PAML program (A = nonsynonymous substitutions per nonsynonymous sites rate (dN); B = synonymous substitutions per nonsynonymous sites rate (dS); C = dN/dS.