Convert Data to IM-Metingen Format: Single Table Setup Database
"Instructions for converting data to the IM-Metingen format, including setup of a single table database. Learn how to customize, split, and map data columns efficiently. Follow steps for transforming a single table into two tables, optimizing data for import into a database. Explore the process of importing and setting up database tables. Get insights on handling different data formats and managing data redundancy. Includes helpful images and detailed instructions."
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
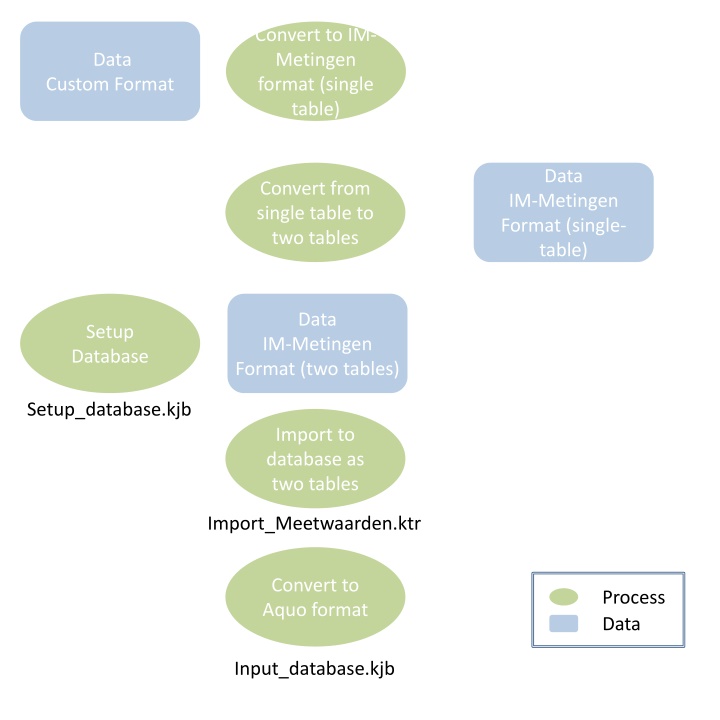
Convert to IM-Metingen format (single table) Convert to IM- Metingen format (single table) Data Custom Format Convert from single table to two tables Data Convert from single table to two tables IM-Metingen Format (single- table) Setup Database Data Setup Database IM-Metingen Format (two tables) Setup_database.kjb Import to database as two tables Import to database as two tables Import_Meetwaarden.ktr Convert to Aquo format Convert to Aquo format Process Data Input_database.kjb
Convert to IM- Metingen format (single table) Run get_data_*.ktr HOME HOME Data Custom format Depending on the type of data/the data format, you can use any of the example data imports that are available in: repos\aquo\Transformation\Get_data\Get_data_and_process_to_IM\ Or repos\aquo\Transformation\Readers\ This contains examples for: CSV, SHAPE, Microsoft Access database, or postgres database. The following steps are generally taken: Read data, split columns, rename columns, map parameters, melt columns (make 1 (or 2) column(s) of multiple columns), rename values, map locations, export meetwaarden and export meetpunten.
Convert from single table to two tables Run get_data_IM_Metingen_*.ktr HOME HOME Data IM Metingen format (single table) In general the data is presented in a single table conform the IM- Metingen uitwissel formaat. To load the data and process it in later steps it is important to already remove some redundancy. The point locations are filtered to be a unique set and stored in a new meetpunten.csv file. The meetwaarden are copied and stored in the meetwaarden.csv file. The following steps are generally taken: Read data, pick unique values, export meetwaarden and export meetpunten.
Import to database as two tables Run Import_Meetwaarden.ktr Or Import_Meetwaarden_extended.ktr HOME HOME Data IM Metingen format (single table) In this step the data is imported into the database. When no polygons and lines are present in the data it is sufficient to use import_meetwaarden.ktr, if there are polygons and lines involved, the IM-Metingen uitwisselformat is extended with two additional columns, which can also be imported with Import_Meetwaarden_extenden.ktr. If any rows are not import correctly those will be shown in the logs. The result is a filled table in the database! Note that these tables should already exist in the database. If not they can be created with the transformation: Create_tables.ktr, which also depends on the schema in which the tables are created, default is dump. (Use create_schema.ktr to create the dump schema.
Setup Database Run Setup_Database.jobs HOME HOME Get data from Domain tables (server) Get data from Domain tables (CSV) Get data from Domain tables (server) Get data from Domain tables (CSV) Split up large tables Split up large tables Fill additional tables Fill additional tables
Get data from Domain tables (CSV) Get data from Domain tables (CSV) Run Load_Domein_tables_csv.kjb HOME HOME Since some of the bigger domain tables were not imported correctly from the webservice they were manually downloaded and imported as csv. - Parameters - Biotaxon - Bemonsteringsapparaat
Get data from Domain tables (server) Get data from Domain tables (server) Run Domeinnamen_AQUO.kjb HOME HOME Load the following domain tables from the webservice - Waardebewerkingsmethode - Hoedanigheid - Compartiment - Eenheid
Split domain tables Split domain tables Run Split_domain_tables.kjb HOME HOME The following domain tables are not present as table in the webservice but are obtained from the parameters table: - Typering - Substance group - Object group - Grootheid - Chemische stof Furthermore there are some tables of which I only could find data in the example database: - Bepaling grens type - Observed property type class result type type - WKR version
Convert to Aquo format Run Import_Aquo_ETL HOME HOME Data IM-Metingen two tables Add to Monitoring_object Add to Monitoring_o bject Add to observedproperty_types Add to observedprop erty_types Add to Observation Add to Observation Add to bio_observati on Add to Sample
Add to Monitoring_object Add to Monitoring_o bject Add to monitoring object HOME HOME Monitoring_object Mno_id Inspire_id_local_id Inspire_id_namespace_id Wkrv_id Monitoring_object_type Name Additional_description Geometry Last_change_time Geometry_etrs89 Begin_lifespan_version End_lifespan_version Primary_geo_column Reason_change Primary_geodetic_reference_id
Add to Observed_property_type Add to Add to observed property type Observed_pro perty_type HOME HOME Observed_property_type opt_id observed_property_type_class result_type hoedanigheid_id object_group_id quantity_id taxa_group_id substance_group_id indicator_id classification_group_id remarks d_begin d_eind last_changed_date d_status
Add to observation Add to observation Add to observation HOME HOME observation obs_id phenomenon_time_begin phenomenon_time_end result_time valid_time_begin valid_time_end observed_property_type quality_indicator_id numeric_value_uom_id value_processing_method_id limit_symbol_id description classified_result_id remarks result_nature_id start_depth end_depth obs_parm_parent_id mno_id integer,
CREATE TABLE public.obs_process_meetapparaat ( obp_id integer NOT NULL, public.observation_process doen we mar_id integer NOT NULL, Zoek uit domains.veldapparaat Doen we last_change_time timestamp without time zone NOT NULL,
CREATE TABLE public.observation_process ( obp_id integer NOT NULL DEFAULT nextval('observation_process_seq'::regclass), ore_id integer, public.organisatorische_eenheid (gekoppelde tabel misschien?> Accreditatiestatus.omschrijving last_change_time timestamp without time zone NOT NULL, wbpm_id integer, public.waarde_bepalings_methode_type Gekoppelde tabel sensing_location_type_id integer, domains.locatietype_waardebepaling ( Doen we sample_pre_treatment_method_id integer, domains.monstervoorbehandeling (Doen sample_destruction_method_id integer, domains.monsterbewerkingsmethode certification_id integer, domains.certificeringscode (Doen we niet) analytical_technique_id integer, domains.waardebepalingstechniek Waardebepalingstechniek.code accreditation_org_id integer, public.organisatorische_eenheid (gekoppelde tabel nen3610_id_namespace_id integer, public.namespace_type Bemonsteringsmethode.code ) Hier vullen we ore_id in als deze beschikbaar is, anders enkel: Last_change_time, waarde bepalings methode als die beschikbaar is en verder niks.
CREATE TABLE public.observation_process ( obp_id integer NOT NULL DEFAULT nextval('observation_process_seq'::regclass), ore_id integer, last_change_time timestamp without time zone NOT NULL, wbpm_id integer, sensing_location_type_id integer, sample_pre_treatment_method_id integer, sample_destruction_method_id integer, certification_id integer, analytical_technique_id integer, accreditation_org_id integer, nen3610_id_namespace_id integer, CONSTRAINT pk_obp PRIMARY KEY (obp_id), CONSTRAINT fk_obp_accreditation_org FOREIGN KEY (accreditation_org_id) REFERENCES public.organisatorische_eenheid (ore_id) MATCH SIMPLE ON UPDATE NO ACTION ON DELETE NO ACTION, CONSTRAINT fk_obp_analytical_technique FOREIGN KEY (analytical_technique_id) REFERENCES domains.waardebepalingstechniek (wbpt_id) MATCH SIMPLE ON UPDATE NO ACTION ON DELETE NO ACTION, CONSTRAINT fk_obp_certification FOREIGN KEY (certification_id) REFERENCES domains.certificeringscode (ctf_id) MATCH SIMPLE ON UPDATE NO ACTION ON DELETE NO ACTION, CONSTRAINT fk_obp_sample_destruction_method FOREIGN KEY (sample_destruction_method_id REFERENCES domains.monsterbewerkingsmethode (mbm_id) MATCH SIMPLE ON UPDATE NO ACTION ON DELETE NO ACTION, CONSTRAINT fk_obp_sample_pre_treatment_method FOREIGN KEY (sample_pre_treatment_method_id REFERENCES domains.monstervoorbehandeling (mvb_id) MATCH SIMPLE