Digital Pathology Image Interpretation: Process, Challenges, and Plans
Explore the world of digital pathology image interpretation presented by Vahid Khalkhali from the Neural Engineering Data Consortium at Temple University. Learn about the current state, technical challenges, and future plans involved in the digital pathology process pipeline. Discover the high-performance computing infrastructure, image digitization, data organization, and anonymization procedures in this cutting-edge field of medical science. Join the journey of leveraging technology to revolutionize disease diagnosis and treatment.
Uploaded on | 3 Views
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
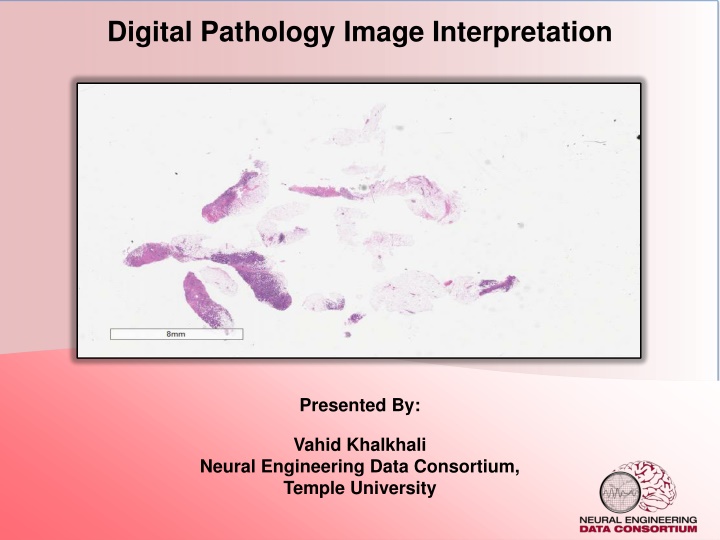
Digital Pathology Image Interpretation Presented By: Vahid Khalkhali Neural Engineering Data Consortium, Temple University
Outline Digital Pathology Process Pipeline Our Current State Technical Challenges Plans Q & A V. Khalkhali: Digital Pathology 1 May 15, 2020
Digital Pathology Pathology is a branch of medical science focused on the cause, origin, and nature, of disease A typical pathology laboratory workflow involves the preparation of a very thin tissue specimen mounted on a glass slide. Digital Pathology refers to the process of digitizing an analog image so that images can be retrieved, manipulated, and evaluated by computers. There are currently two dominant vendors in the commercial marketplace. Our group use Leica Biosystems Aperio. The slide files are 3 or 4 flat 2d images. V. Khalkhali: Digital Pathology 2 May 15, 2020
Process Pipeline Neuronix: high-performance computing (HPC) cluster Computing Infrastructure Aperio AT2 high volume scanner (JPEG2000) Image Digitization Aperio eSM Web Server with naming conventions Data Organization Deidentification and transfer to Neuronix Data Anonymization Freehand annotation, XML labels Data Annotation V. Khalkhali: Digital Pathology 3 May 15, 2020
Our Current State Currently, our group has done all the fundamental steps. Organizing the overall process is needed. Neuronix: high-performance computing (HPC) cluster Computing Infrastructure Aperio AT2 high volume scanner (JPEG2000) Image Digitization Aperio eSM Web Server with naming conventions Data Organization Deidentification and transfer to Neuronix Data Anonymization Freehand annotation, XML labels Data Annotation V. Khalkhali: Digital Pathology 4 May 15, 2020
Technical Challenges Anonymization procedure and tools need to be documented. It seems that the group has not decided about the labels container. The large size of images in the lowest level of slides makes most computations infeasible. If the number of images increases significantly, the storage space can be a challenge. Automated cancer recognition cannot be built on low number of annotated data. Speeding up the speed of annotation and training more annotators are necessary. V. Khalkhali: Digital Pathology 5 May 15, 2020
Plan Near term: Finish the deidentification process with documents Decide about the labels container and make necessary conversions Decide about images data manipulation Long term: Generating 100,000 images. Speed up annotation process Making automated cancer recognition system V. Khalkhali: Digital Pathology 6 May 15, 2020
Q & A Thank You! V. Khalkhali: Digital Pathology 7 May 15, 2020
References Shawki, N., Shadhin, M. G. M., Elseify, T., Jakielaszek, L., Farkas, T., Persidsky, Y., Picone, J. (2020). The Temple University Digital Pathology Corpus. In I. Obeid, I. Selesnick, & J. Picone (Eds.), Signal Processing in Medicine and Biology: Emerging Trends in Research and Applications (1st ed., pp. 67 104). New York City, New York, USA: Springer. https://doi.org/10.1007/978-3-030- 36844-9 Aperio Digital Pathology Solutions, Accessed on: May. 14, 2020, [Online]. Available: https://www2.leicabiosystems.com/ V. Khalkhali: Digital Pathology 8 May 15, 2020