DNA Methylation
Delve into the world of DNA methylation, exploring its quantification, experimental methods, and data analysis techniques. From understanding methylated DNA to aligning sequences and calculating conserved cytosine percentages, this content provides a comprehensive overview of this essential biological process.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
DNA Methylation Kat Kessler, James Vesto
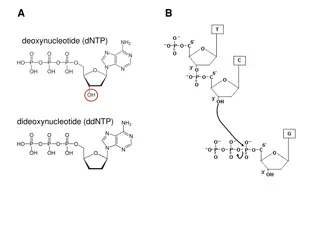
DNA Methylation Quantification What is DNA Methylation? How do we know if we have methylated DNA? Why do we care?
Data Sanger sequence? Simulate toy sequences by randomly changing C T Keep track of how many are changed/conserved
Main( ) original String of Known experimental String of DNA Toy Data Orientation, Original Align, Experimental Align Align main(original, experimental) Methylated count, Possible count, CPG Methylated Count, CPG Possible count methyl(Original alignment, Experimental alignment) Percent score,Percent CPG score Methylated Count) Percent methyl(Possible Count, Methylated Count, CPG Possible Count, CPG return
Align main (original, experimental) Indel -1 Match +1 Mismatch -1 Score, Ori Align, Exp Align Global Align (Original, Experimental, Indel, Match, Mismatch) if Lagging Score > Lead Score or (Lag Score < Lead Score) Orientation "Lag or "Lead" Align Original Lag Ori Align or Lead Ori Align Align Lag Exp Align or Lead Exp Align return Orientation, Original Align, Experimental Align
Methyl(Original, Experimental, Orientation) if Orientation = lead or lag nucleotide C or G CPG G or C for i 0 to |original| if original[i] = nucleotide Possible Count Possible Count + 1 if experimental[i] = nucleotide Methylated Count Methylated Count + 1 if i>0 and original[i-1] or [i+1] = CPG CPG Possible Count CPG Possible Count+1 if experimental[i]= nucleotide CPG Methylated Count CPG Methylated Count + 1 return Methylated count, Possible count, CPG Methylated Count, CPG Possible Count
Obtaining Percentage Of Conserved Cytosine Create alignment keep track of C and CpG GlobalAlign LCS Indels, matches, mismatches? Forward/reverse? Conserved C / Total C CpG complexes? Conserved CpG / Total CpG How to include in results
Results Both forward and reverse alignments and their LCS Run forward or reverse methylation function conserved nucleotides! C T G A Nucleotide counts Overall percent methylation CpG methylation Toy data had 65 possible sites of methylation , 41 conserved nucleotides, obtained 53.846% methylation, expected 63.07% methylation Molecular information!