Enhancing Genomic Analysis Through Collaborative Data Sharing
Explore the collaborative efforts between NHGRI, CCDG, and TOPMed in jointly calling variant data to improve genotype quality and facilitate further analysis. Proposed use cases include enhancing variant servers, imputation reference panels, and enabling population genetic studies. Ongoing efforts focus on permissions and data distribution for various downstream applications.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
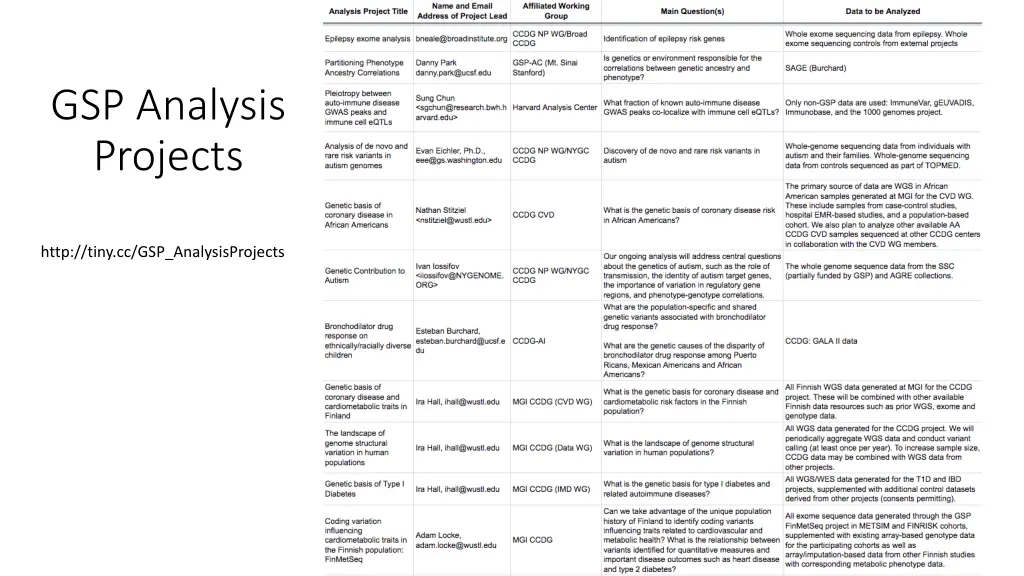
GSP Analysis Projects http://tiny.cc/GSP_AnalysisProjects
GSP-funded Presentations at ASHG 2017 http://gsp-hg.org/gsp-funded-presentations
Goals of NHGRI CCDG-NHLBI TOPMed Joint Calling Currently, TOPMed has ~65k WGS samples sequenced, CCDG has ~22k (ultimately, ~120k and ~85k, respectively). Jointly calling all the samples together would improve the overall quality of the calls and provide a large dataset of harmonized genotypes for further analysis. Forging mechanisms for such data sharing across NIH programs will help advance the best science, increased sample sizes and ethnic diversity. This joint calling exercise will allow the identification of computational issues, refinement and enhancement of pipelines, and assessment of the relative merits of different calling algorithms to develop best practices for different variant types.
Proposed Use-cases for the Jointly-called Data Enhance the information in variant servers (e.g. Bravo, gnomAD) Enhance information for cosmopolitan imputation reference panels Facilitate functional annotation Develop large samples to serve as common controls Enable population genetic studies Enable yet larger studies of genotype:phenotype association. Red indicates initial use case; others will follow after detailed discussions related to consent and controlled access.
CCDG studies TOPMed studies TOPMed Seq Ctrs CCDG Seq Ctrs TOPMed IRC Amazon Google 5 Calling Groups Variant Server CCDG vcfs CCDG CC pVCFs dbGaP TOPMed Ex Area Imputation server TOPMed vcfs Haplotype data
Ongoing efforts 1. Permission from studies for CCDG and TOPMed CRAMs to have variants jointly called. 2. Permission for distribution of variant summary data to Variant and Imputation Servers. 3. Distribution of gVCFs to TOPMed, CCDG and dbGaP (for public release) can be handled in the same way as currently done for program- specific data distribution; no new permissions are needed. 4. Permissions for other downstream use-cases (common controls, population genetics, genotype-phenotype association studies) may be via the usual DAC mechanism or a new mechanism TBD.
