Enzyme Catalysis and Classification
Enzymes are biological polymers that catalyze chemical reactions, facilitating the conversion of substrates into products. Their catalytic activity relies on the integrity of their protein conformation. Enzymes are highly selective catalysts, specific to the type of reaction and substrates involved. The uniqueness of their active sites makes them either substrate-specific or group-specific, influencing their recognition and cleavage patterns. Additionally, enzymes exhibit substrate specificity, stereo specificity, and essential protein structures that ensure their catalytic activity.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
BIOCHEMISTY Course No.-DTC-111, Credit Hours 2 (1+1) ENZYME CATALYSIS AND CLASSIFICATION BINITA RANI ASSOCIATE PROFESSOR (DAIRY CHEMISTRY) FACULTY OF DAIRY TECHNOLOGY S.G.I.D.T., BVC CAMPUS, P.O.- BVC, DIST.-PATNA-800014
Enzymes are biologic polymers => catalyze chemical reactions. majority of enzymes => proteins (exception => few catalytic RNA molecules, or ribozymes) . enzymes catalyze => conversion of one or more compounds (substrates) => one or more different compounds (products). enhance the rates of corresponding non-catalyzed reaction. Catalysts => do not affect reaction equilibria.
enzymes => neither consumed nor permanently altered => as a consequence of their participation in a reaction. Their catalytic activity depends => integrity of => native protein conformation. If enzyme => denatured or dissociated => its subunits => catalytic activity is usually lost. If enzyme => broken down => component amino acids => catalytic activity => destroyed. primary, secondary, tertiary, and quaternary structures => of protein enzymes => essential => catalytic activity.
Enzymes => extremely selective catalysts enzymes => specific both for type of reaction catalyzed and for a single substrate or a small set of closely related substrates Enzymes are substrate specific or group specific
Conformation => complex proteins and uniqueness of active site => of enzymes make them substrate specific or absolute group specific. glucokinase recognize glucose => as absolute substrate while Hexokinase recognize => aldohexose (Glucose / mannose etc.) => substrate. trypsin, chymotrypsin and elastase => cleaves proteins on carboxyl side of positively charged (Lysine, Arginine), aromatic amino acids (Tyrosine, Phenylalanine) and small group side chains (alanine, glycine) amino acids, respectively.
Enzymes => stereospecific catalysts typically catalyze reactions only of specific stereoisomers of a given compound, e.g., D- but not L-sugars L- but not D-amino acids
Enzymes show geometric specificity Enzyme bind substrates through at least three points of attachment => can even convert nonchiral substrates to chiral products.
Common features of enzyme active site active site of an enzyme => generally a pocket or cleft => specialized => recognize specific substrates => and catalyze chemical transformations. Hence => enzyme and substrate => have complementary shapes. active site => small part of => total volume of enzyme. It is formed in => three-dimensional structure => by a collection of different amino acids (active-site residues) => that may or may not be adjacent in => primary sequence.
interactions between => active site and the substrate occur via => same forces that => stabilize protein structure: hydrophobic interactions, electrostatic interactions (charge charge), hydrogen bonding, and Vander Waals interactions. active sites => do not simply bind substrates, they also provide => catalytic groups => facilitate the chemistry and provide specific interactions => that stabilize formation of transition state for => chemical reaction.
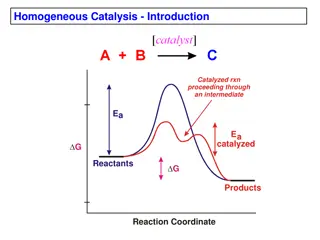
During chemical reaction => structure of substrate changes => structure of product. Somewhere in b/w => some bonds are partly broken => others are partly formed. Transition state => highest- energy arrangement of atoms => that is intermediate in structure b/w structure of reactants and structure of products. diagram shows => free energy of the reactants, transition state, and product.
free-energy difference b/w product and reactant is => free- energychange for => overall reaction. free-energy change tells => how favorable the reaction is thermodynamically => not about its rate. Some energy must be put => into the reactants before => they can be converted => products. This activation energy provides => barrier to the reaction the higher the barrier => the slower the reaction. difference in free energy b/w transition state and reactant(s) => free energy of activation.
Reaction rates => increased by => raising the temperature => thereby increasing the number of molecules with sufficient energy => to overcome the energy barrier. the activation energy => lowered by adding a catalyst (Fig.). Catalysts => enhance rk rates by => lowering activation energies. role of enzymes is => accelerate the interconversion of S and P. enzyme is not used up in process, and => equilibrium point is unaffected. reaction reaches equilibrium much faster => when appropriate enzyme is present => because rate of reaction is increased.
Enzyme Classification enzymes => named by adding suffix -ase => name of their substrate or to a word or phrase describing their activity => urease catalyzes hydrolysis of urea, and DNA polymerase => polymerization of nucleotides to form DNA. Some enzymes => named => for a broad function => before specific reaction catalyzed was known => lysozyme was named => its ability to lyse bacterial cell walls. Sometimes same enzyme => two or more names, or two different enzymes => same name. Because of such ambiguities => biochemists, by international agreement => have adopted a system => naming and classifying enzymes.
This system => divides enzymes => six classes => each with subclasses => based on the type of reaction catalyzed . Each enzyme is assigned => four-part classification number and a systematic name => which identifies the reaction it catalyzes , e.g., formal systematic name of enzyme catalyzing the reaction is ATP: glucose phosphotransferase => which indicates => it catalyzes the transfer of a phosphoryl group from ATP to glucose.
Its Enzyme Commission number ( E. C. number) is 2.7.1.1. first number (2) => major class name (transferase); second number (7) => subclass (phosphotransferase); third number => sub-subclass (1) => a phosphotransferase with a hydroxyl group as acceptor; and fourth number (1) => enzyme number in the sub-subclass.
Cofactors Enzymes => molecular weights ranging => 12,000 to > 1 million. Some enzymes require => no chemical groups for activity => other than => their amino acid residues. Others require => additional chemical component => cofactor => either one or more inorganic ions => Fe2+, Mg2+, Mn2+, or Zn2+, / a complex organic / metallo-organic molecule => coenzyme. Some enzymes require => both a coenzyme and one or more metal ions => activity.
A coenzyme or metal ion => very tightly or even covalently bound => enzyme protein => a prosthetic group. A complete, catalytically active enzyme => together with its bound coenzyme and/or metal ions => holoenzyme. protein part of such an enzyme => apoenzyme or apoprotein. Coenzymes act => transient carriers of specific functional groups. Most are derived from => vitamins, organic nutrients required => in small amounts in the diet.
Coenzymes as Transient Carriers of Specific Atoms or Functional Group