Generate Clusters of Identical Fragments on Glass Surface
Learn about the process of generating clusters of identical fragments on a glass surface using solid-phase PCR amplification. Understand how clusters are created, the use of forward and reverse primers, polymerase, and nucleotides, and the impact of temperature changes. Discover the bridge-break PCR amplification method to produce distinct clusters without overlap, leading to the successful generation of local clusters from single template molecules.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
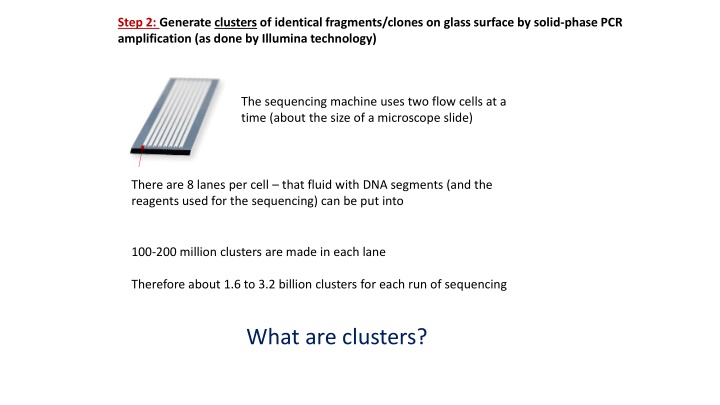
Step 2: Generate clusters of identical fragments/clones on glass surface by solid-phase PCR amplification (as done by Illumina technology) The sequencing machine uses two flow cells at a time (about the size of a microscope slide) There are 8 lanes per cell that fluid with DNA segments (and the reagents used for the sequencing) can be put into 100-200 million clusters are made in each lane Therefore about 1.6 to 3.2 billion clusters for each run of sequencing What are clusters?
Step 2: Generate clusters of identical fragments/clones on glass surface by solid-phase PCR amplification (as done by Illumina technology) Forward and reverse primers are covalently attached to the slide at high-density- in no particular pattern These match the adaptors at the ends of the DNA segments that have been produced DNA is added to the slides and allowed to anneal to the immobilized primers diluted to keep the sites well separated so when we amplify to form the clusters, each one is far from the others
What if we add polymerase and nucleotides (and the right buffers, etc.)? So now we have this: C C F F R R
Now what if we increase the temperature? C F R F R Template is released and washed away
Step 2: Generate clusters of identical fragments/clones on glass surface by solid-phase PCR amplification (as done by Illumina technology) ****Clusters are distinct, separated in space (no overlap) Bridge-break PCR amplification generates local clusters from single template molecules. So you start with just one molecule bound, over a series of amplification cycles, you get clusters of forward and reverse strands of the same sequence How are the clusters made?
Next Steps: Bridge amplification- generate the clusters of the same sequence
Annealing 60C F F R R F F R R MANY CYCLES Like this 72C Denaturing 95C F F R R F F R R
So, where are we so far? We have in each lane 100 200 million spots or clusters, each of which represents one small fragment of DNA The fragments are 100-200 nucleotides in length Now we want to sequence the fragments