Genomic Prediction Overview: Simplified Process Map for Breeding
Discover how genomic prediction in breeding is streamlined through a simplified process map, covering SOPs, checklists, and key metrics. Learn about plan prediction, genotype/phenotype acquisition, secondary QC, and more.
Uploaded on | 1 Views
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
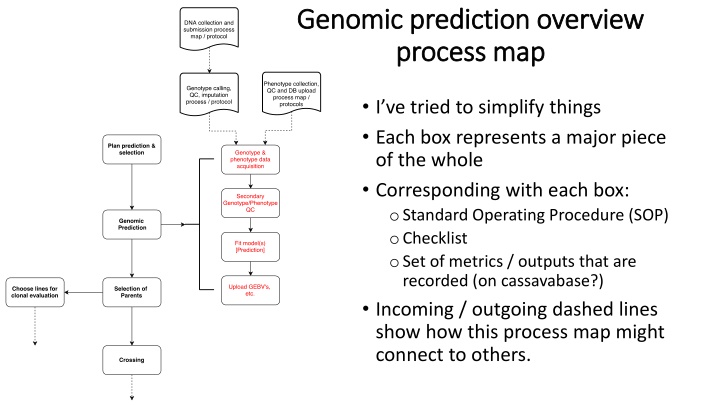
Genomic prediction overview Genomic prediction overview process map process map I ve tried to simplify things Each box represents a major piece of the whole Corresponding with each box: oStandard Operating Procedure (SOP) oChecklist oSet of metrics / outputs that are recorded (on cassavabase?) Incoming / outgoing dashed lines show how this process map might connect to others.
SOP / Checklist Plan prediction Plan prediction & selection & selection 1. Breeder and analyst conference Meeting to discuss upcoming prediction. What is needed, when it is needed, etc. 2. Analysis leverages either pre-existing information and/or does new analysis to decide on the best approach for the upcoming prediction. This means testing different training populations, prediction models, etc. using, for example, cross-validation or cross-generation prediction within the available training data. 3. Breeder and analyst conference #2 In some circumstances, this might not be necessary. Basically, consultation should be done to get approval for proposed approach. Outputs / Metrics / Tracking Due date for prediction Traits / Selection index to be used Target population of environments (TPOE) Selection candidates to be predicted Prediction models to be used Record / report on analyses done and data leveraged forming the basis for decisions made
Genomic prediction Genomic prediction Genotype / Phenotype Genotype / Phenotype Acquisition Acquisition SOP / Checklist 1. Phenotype data accessed from Cassavabase 2. SNP data either accessed from DB (Cassavabase / GOBII) or a sample list suitable for subsetting external datasets is accessed. 3. Review meta-data / records of previous QC and filtration done on data Outputs / Metrics / Tracking Time and parameters of DB download (sufficient to recreate) Checklist mark that review of prior QC completed with any relevant comments
SOP / Checklist Genomic prediction Genomic prediction Secondary QC Secondary QC 1. Matching of DNA records with phenotypic records Presently, this is a critical, non-trivial step In the future, the DB can handle this and it will become trivial 2. Genotype QC and preparation Formatting, subsetting, filtration (e.g. minor allele frequency, imputation quality) Construction of genomic relationship (aka Kinship) matrix 3. Phenotype QC and preparation Probably a long and involved SOP. Steps include: Check for typos (e.g. based on columns that should be readable as numeric but that R treats as character) Check for impossible / improbable values Cross-check number sprouting with number harvested Cross-check record of harvest (number stands) with presence of harvest data (root yield, dry matter, brown streak). Examine trait distributions / correlations Fit mixed-model and obtain BLUPs (correction of data for trial design). Only applies if two- step genomic prediction to be used. Outputs / Metrics / Tracking Record of DNA-Phenotype matching chosen Record of filters applied, SNPs removed/kept Record / checklist marks of phenotype data QC done. What was removed or altered and in what way? Relevant comments on process. Code used. BLUPs, reliabilities, weights and relevant model outputs from first of two-step prediction procedure (when applicable).
Genomic prediction Genomic prediction Fit model(s) Fit model(s) [make prediction] [make prediction] SOP / Checklist 1. Fit genomic prediction models 2. Review results Variance components Heritability Distribution of GEBV and reliabilities Compare to previous results (for clones that have been predicted before) 3. Format outputs and report 4. Upload to database and notify breeder(s). Outputs / Metrics / Tracking Spreadsheet with predicted GEBV, etc. Table(s) summarizing models fit (variance components, heritability, etc.) Record / Report explaining what was done in a manner suitable to enable results to be reproduced by another scientist. Code.
SOP / Checklist 1. Breeder(s) download predictions and associated outputs (e.g. corresponding report and model summary) 2. Review and (hopefully) approve results Questions for and further analysis by the analyst, if necessary, here. 3. Standardize GEBV and create selection index 4. Selection Selection of Parents Selection of Parents Kept this simple as its not my area really. Potentially, cassavabase could be used to display useful information from the prediction that was uploaded in the previous step. SolGS already has useful outputs for its predictions. Outputs / Metrics / Tracking Record of selection weights and index. Record of clones select parents. Expected selection intensity. Expected response to selection.










