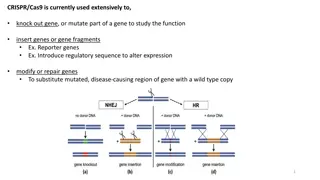
Germline Variant Analysis and De Novo Results in Cancer Subgroup Study
Explore the MOAT Germline Variant Analysis and its application in identifying de novo mutations in cancer subgroups. Discover the significant genes and sensitivity/specificity trade-offs for better understanding genetic variants. Learn about ANNOVAR for functional annotation of genetic variants.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
MOAT Germline Variant Analysis Lucas Lochovsky A Cancer subgroup 2016-11-04 1
Relevant MOAT Parameters MOAT-a d_min: Minimum distance from annotation for picking a random bin d_max: Maximum distance from annotation for picking a random bin MOAT-v bin width: The width that represents the local genome context. Variants are shuffled to new locations within their containing bin. min bin width: In the event that an especially small bin is formed, either due to a chromosome end or subtraction of a blacklist region, merge the bin with the nearest full size bin if it s below this width. 3
MOAT Germline Variant Analysis Demonstrate MOAT capability on germline variants Work with a large (~5000) de novo mutation set from: Kong, A. et al.Rate of de novo mutations and the importance of father s age to disease risk. Nature488, 471 475 (2012). Sequencing of 78 trios, 44 of which have autism spectrum disorder (ASD) Evaluate MOAT recapitulation of ASD-associated genes 4
De novo results Results are typically a range of p-values, but after BH correction, all p- values are mapped to either 0s or 1s Counts for p-value=0 genes: MOAT-a Significant Gene Count MOAT-v Significant Gene Count 140 140 120 120 100 100 # signif genes # signif genes 80 80 60 60 40 40 20 20 0 0 10kb 40kb 100kb 1mb 100kb 10kb 1kb d_max Bin width 5
De novo results Kong et al. drew attention to variants in the exons of 3 ASD- associated genes NRXN1, CUL3, EPHB2 MOAT-v with low bin width produces a small set But at 1kb/10kb, only CUL3 is recapitulated At higher bin widths, all 3 are recapitulated, but we also see many more unrelated genes Sensitivity/specificity tradeoff 6
De novo results with ANNOVAR Combine recurrence with functional impact For gene-based annotations, there is ANNOVAR Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data Nucleic Acids Research, 38:e164, 2010 Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web Journal of Medical Genetics, 49:433-436, 2012 Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR Nature Protocols, 10:1556-1566, 2015 Pick out the nonsynonymous variants 7
De novo results Analysis? # Analysis 1 ANNOVAR? applied? after? MOAT-v 2 Only? ANNOVAR 3 Only? MOAT-v #? Signif? Genes #? autism-related #? neurodisease-related #? unrelated 31 18 62 27 65 23 Influence of Analysis Type on Percent of Related Genes (Autism + Neurodegenerative) 4 7 6 9 28 36 80% 71% 70% 60% 55% 50% Percent related 45% 40% 30% 20% 10% 0% 8 1 2 3 Analysis Type