Identification of an Acetyl Esterase in Bacillus sp. HR21-6 Supernant
This study focuses on identifying an acetyl esterase in the supernatant of the environmental strain Bacillus sp. HR21-6. The research involves the purification and characterization of the enzyme, as well as molecular analysis and comparison with related members of the SGNH subfamily. Various experimental methods and findings are discussed, shedding light on the functional properties and structural features of the acetyl esterase AE6L.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
Identification of an acetyl esterase in the supernant of the environmental strain Bacillus sp. HR21-6 Leyre S nchez-Barrionuevoa,b, Jes s Mateosc1, Patricia Fern ndez- Puentec, Paloma Beginesd, Jos G. Fern ndez-Bola osd, Gabriel Guti rreza, David C novasa*, Encarnaci n Melladob* aDepartment of Genetics, Faculty of Biology, University of Seville, Seville, Spain bDepartment of Microbiology and Parasitology, Faculty of Pharmacy, University of Seville, Seville, Spain cGrupo Investigaci n Biom dica de A Coru a (INIBIC), Complexo Hospitalario Universitario de A Coru a (CHUAC-SERGAS). A Coru a, Spain de Prote mica-PBR2-ProteoRed/ISCIII. Instituto de dDepartment of Organic Chemistry, Faculty of Chemistry, University of Seville, Seville, Spain 1Pressent address. Marine Research Institute-Spanish National Research Council (CSIC), Vigo, Spain *Corresponding author: Encarnaci n Mellado: emellado@us.es David C novas: davidc@us.es
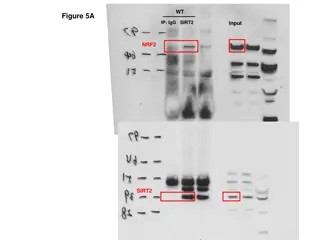
Peptide GATPLLLTPVGR charge 3+ Replicate 1 Replicate 2 FSEP OMVs Fig. S1. MRM analysis of the peptides GATPLLLTPVGR (charge +3) and YTAQGVSVDNR (charge +3 and +2) of the acetyl esterase AE6L detected in the FSEP fraction of the supernatant of Bacillus sp. HR21-6.
Replicate 1 Replicate 2 Peptide YTAQGVSVDNR charge 3+ FSEP OMVs Peptide YTAQGVSVDNR charge 2+ FSEP OMVs Supplementary Figure 1 (cont.).
30C 20 C A 1 2 - M - 4 1 2 3 4 M 3 70 kDa 40 kDa 25 kDa B - + IF M SF 70 kDa 40 kDa 25 kDa Fig. S2. SDS-PAGE analysis ofAE6L expressed in E. coli BL21(DE3). (A) Expression ofAE6L in E. coli BL21(DE3) at two different temperatures (30 C and 20 C) for 4 h. (B) Identification of AE6L in the soluble fraction (SF) of the E. coli BL21(DE3) pMAB-AE6L induced with 1 mM salicilate at 30 C. M, molecular mass marker. -, BL21 (DE3) pMAB-AE6L without inducing, +, BL21 (DE3) pMAB-AE6L induced with 1 mM salycilate at 30 C; 1, BL21 (DE3) pMAB-AE6L with 0.2 mM salicylate; 2, BL21 (DE3) pMAB-AE6L with 1 mM salicylate; 3, pMAB-AE6L with 1 mM salicylate + 3-methylbenzoate.; 4, pMAB- AE6L with 1 mM of IPTG; IF, insoluble fraction; SF, soluble fraction. Polyacrylamide gels (12% w/v) were stained with Coomassie brilliant blue. AE6L protein, indicated by the arrow, was identified according to the estimated mass from SDS PAGE compared with the estimated mass from the aminoacid sequence.
Block I Block II Block III Block V Fig. S3. Multiple aminoacid sequence alignments of the AE6L from Bacillus sp. HR21-6 with related members of the SGNH subfamily. Rhamnogalacturonan acetylesterase proteins from Bacillus subtilis ATCC 6633 (O31523.1), Rhamnogalacturonan acetylesterase from Bacillus subtilis subsp. subtilis str. 168 (O31528.1), Rhamnogalacturonan acetylesterase Rhamnogalacturonan acetylesterase from Aspergillus aculeatus (Q0017.1), Rhamnogalacturonan acetylesterase from Apergillus aculeatus (1DEO_A), hypothetical protein YxiM from Bacillus subtilis (2O14_A), Rhamnogalacturonan acetylesterase from Bacillus halodurans C-125 (BAB04834.1). Four conserved sequence blocks (blocks I-V) found in all SGNH hydrolases are boxed. Identical and similar residues are indicated by asterisks and dots below the sequence. Alignment was obtained from the T-COFFE v.1 program. Selected residues related to the catalytic triad are indicated in blue, the residues described as related to the substrate specificity are indicated in yellow. Two other residues are highlighted in green in the AE6L sequence due to the significant differences found after the alignment analysis. from (O32563), Dickeya chrysanthemi
Table S1. Proteins detected in the FSEP and OMV fractions of the exoproteome of Bacillus sp. HR21-6 FSEP 48 h Peptides (95%) Protein 1 2 3 4 FIG01248074: hypothetical protein Catalase (EC 1.11.1.6) FIG01246635: hypothetical protein 24 8 7 6 Dihydrolipoamide dehydrogenase of pyruvate dehydrogenase complex (EC 1.8.1.4) Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein OppA (TC 3.A.1.5.1) serine alkaline protease (subtilisin E)( EC:3.4.21.62 ) Phosphopentomutase (EC 5.4.2.7) ATP carboxykinase [ATP] (EC 4.1.1.49) oxidoreductase of aldo/keto reductase family, subgroup 1 glutamyl endopeptidase precursor Ribonuclease (EC 3.1.27.-) (RNase Bci) rhamnogalacturonan acetylesterase Cell wall surface anchor family protein, LPXTG motif 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (EC 5.4.2.1) Arginase (EC 3.5.3.1) FIG01232504: hypothetical protein Leucine dehydrogenase (EC 1.4.1.9) Alanine dehydrogenase (EC 1.4.1.1) Cytosol aminopeptidase PepA (EC 3.4.11.1) Tellurium resistance protein TerD subtilisin Carlsberg( EC:3.4.21.62 ) Manganese superoxide dismutase (EC 1.15.1.1) FIG01232467: hypothetical protein Methionine synthase II (cobalamin-independent) Lipoprotein yhcN precursor Non-specific DNA-binding protein Dps / Iron-binding ferritin-like antioxidant protein / Ferroxidase (EC 1.16.3.1) Peptidase, M42 family Tripeptide aminopeptidase (EC 3.4.11.4) Phosphate acetyltransferase (EC 2.3.1.8) Oligoendopeptidase F intracellular proteinase inhibitor BsuPI Cysteine synthase (EC 2.5.1.47) Peptidase T (EC 3.4.11.4) NPQTN cell wall anchored protein IsdC ThiJ/PfpI family protein FIG01242571: hypothetical protein Ribosome recycling factor 4-oxalocrotonate tautomerase (EC 5.3.2.-) Gamma-glutamyltranspeptidase (EC 2.3.2.2) NADH-dependent butanol dehydrogenase A (EC 1.1.1.-) NADH-dependent butanol dehydrogenase A (EC 1.1.1.-) (3R)-hydroxymyristoyl-[acyl carrier protein] dehydratase (EC 4.2.1.-) Catalyzes the cleavage of p-aminobenzoyl-glutamate to p-aminobenzoate and glutamate, subunit A Tellurium resistance protein TerD ATP-dependent protease La (EC 3.4.21.53) LonB Type I putative cytochrome P450 hydroxylase GTP cyclohydrolase I (EC 3.5.4.16) type 1 Manganese ABC transporter, periplasmic-binding protein SitA hypothetical protein 5 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 5 3 3 2 2 2 3 3 3 2 2 1 1 1 1 1 1 1 1 1 26 1 27 28 29 30 31 32 33 34 35 36 37 38 39 40 40 41 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 42 1 43 44 45 46 47 48 0 0 1 0 0 0
Table S1 (Cont.) OMV 48 h 1 2 3 Peptides (95%) 20 14 15 Protein Catalase (EC 1.11.1.6) FIG01246635: hypothetical protein Glutamine synthetase type I (EC 6.3.1.2) Dihydrolipoamide dehydrogenase of pyruvate dehydrogenase complex (EC 1.8.1.4) Peptidase, M42 family 6,7-dimethyl-8-ribityllumazine synthase (EC 2.5.1.78) Transaldolase (EC 2.2.1.2) Arginase (EC 3.5.3.1) extracellular serine protease Non-specific DNA-binding protein Dps / Iron-binding ferritin-like antioxidant protein / Ferroxidase (EC 1.16.3.1) Manganese superoxide dismutase (EC 1.15.1.1) FIG01248074: hypothetical protein Leucine dehydrogenase (EC 1.4.1.9) Deblocking aminopeptidase (EC 3.4.11.-) FIG01242571: hypothetical protein 4 8 5 6 7 8 9 6 6 4 4 4 10 4 11 12 13 14 15 3 3 3 2 2 16 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (EC 5.4.2.1) 2 17 18 19 20 21 22 23 24 25 Manganese ABC transporter, periplasmic-binding protein SitA Oligoendopeptidase F ATP carboxykinase [ATP] (EC 4.1.1.49) Manganese ABC transporter, ATP-binding protein SitB cephalosporin C deacetylase Preprotein translocase subunit YajC (TC 3.A.5.1.1) Phosphopentomutase (EC 5.4.2.7) Cytosol aminopeptidase PepA (EC 3.4.11.1) Glutamyl aminopeptidase (EC 3.4.11.7); Deblocking aminopeptidase" Siderophore staphylobactin ABC transporter, substrate-binding protein SirA, putative @ ABC-type Fe3+-siderophore transport system, periplasmic iron- binding component Dihydroneopterin aldolase (EC 4.1.2.25) 3-dehydroquinate dehydratase II (EC 4.2.1.10) 1 1 1 2 1 1 2 1 0 26 0 27 28 0 0
Table S2. Sequence identity of AE6L with most related characterized lipases from different microorganisms Accession number protein % Identity with AE6L (query cover) Species Bacillus stratosphericus LAMA 585 EMI14919.1 100% Bacillus sp. K91 AIZ00775.1 62% (query cover 91%) Bacillus halodurans C-125 BAB06525.1 41% (query cover 87%) Bacillus subtilis O31523.1 38% (query cover 91%) Dickeya chrysanthemi O32563 26% (query cover 90%) Aspergillus aculeatus CAA61858.1 27% (query cover 86%)
Table S3. Primer sequences used for the RT-PCR Primer 5 -Sequence-3 Gen AATCGAAAAAGGCGAAT GGA TCCGTTTGGAAATCCCTT GTT AGAATATCTTGGAGCCGC ATGA TAGTGTACGTTCGCCTTC TGGAT GTATCGATTTGCTCGGTT TCG TCCCAGATGCTCGACAA CAG gyrA122F Gyrase Subunit A gyrA122R rpoB122F DNA polymerase Subunit rpoB122R RT-RGAEF AE6L acetyl esterase RT-RGAER
Table S4. Primer sequences used for theAE6L mutagenesis Sequences in lower case indicate the sites of mutations Primer S9AF S9AR G42AF G42AR D74A D74AR D192AF D192AR H195AF H195AR S98AF S98AR E26QF E26QR T194AF T194AR T10AF T10AR 5 -Sequence-3 GGCAGGCGATgcaACCGTTAGCAG AGATAAATGGTGGTCATG TGCAATTGGTgcaCGTAGCAGCA CGATTATCAACGCTAACAC GATGGGTCATgcaGATGCAAGCACC TGGGTAAACAGCCAATCAC CGGCATTGAAgcaCGTACCCATTTTAC GTGCTCAGCATAAAATAGC AGATCGTACCgcaTTTACCAAAGA TCAATGCCGGTGCTCAGC CCGTCTGGTTgcaCTGGAACTGAAAGA TCTGG GCAATTGCGTCTGCACCT TGGTTGGGGTcaaTTTCTGCATC CCCTGAACCGGACGATCC TGAAGATCGTgcaCATTTTACCAAAGAAG ATGCCGGTGCTCAGCATA AGGCGATAGCgcaGTTAGCAGCT GCCAGATAAATGGTGGTCATG