Information on MTDFNRM and Related Programs
This content provides details on MTDFNRM and its associated programs like MTDFRUN, MTDFPREP, and more. It covers topics such as pedigree files, data preparation, output files, compilation and linking of FORTRAN code, optimization switches, and main programs with subroutines. Additionally, it includes images for better understanding of the processes involved in these programs.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
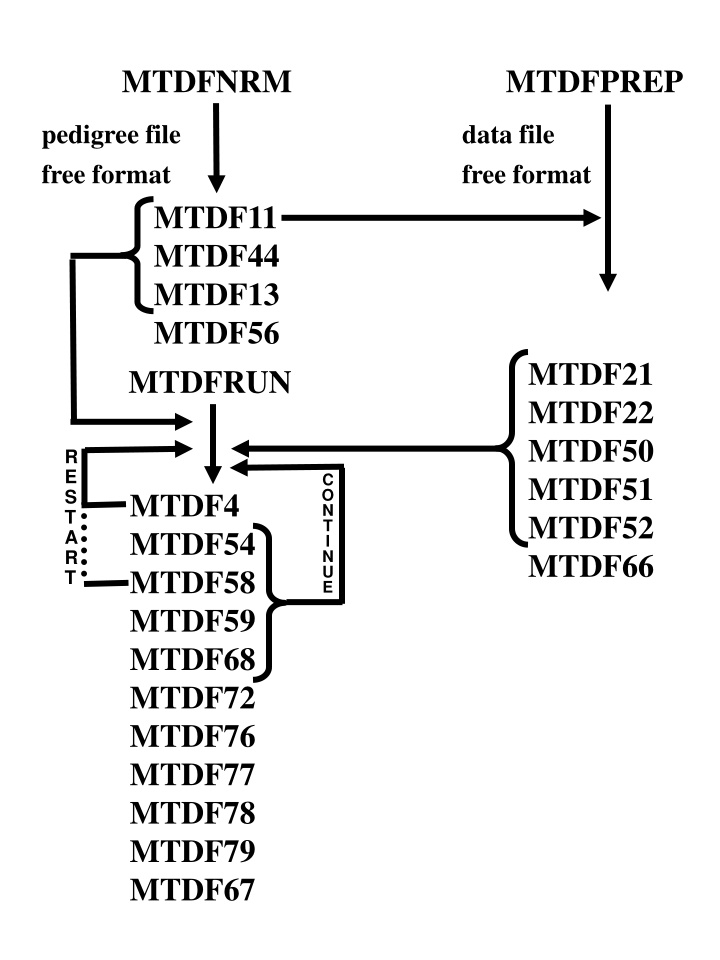
MTDFNRM MTDFPREP pedigree file free format data file free format MTDF11 MTDF44 MTDF13 MTDF56 MTDFRUN MTDF21 MTDF22 MTDF50 MTDF51 MTDF52 MTDF66 R E S T A R T C O N T I N U E MTDF4 MTDF54 MTDF58 MTDF59 MTDF68 MTDF72 MTDF76 MTDF77 MTDF78 MTDF79 MTDF67
MTDFREML Output Files (* = optional) MTDFNRM.EXE MTDFPREP.EXE MTDF11 * MTDF13 MTDF44 MTDF56 No. animals, Recoded and original animal ID No. animals, Recoded and original ID and F for animal, sire, dam BINARY, elements of A-1 Summary for MTDFNRM * MTDF21 * MTDF22 Levels of fixed factors and description Levels of other random factors and description MTDF50 MTDF51 MTDF52 MTDF66 Markers for setting up MME Binary Basically recoded data file Binary for setting up MME Summary for MTDFPREP
MTDFRUN.EXE MTDF4 MTDF54 MTDF58 MTDF59 MTDF68 * MTDF72 MTDF76 * MTDF77 * MTDF78 * MTDF79 * MTDF67 Updated start file Current simplex, -2 logL and VC Reorder file for SPARSPAK File of user imposed constraints History of -2 logL and VC F, PBV, SEP and accuracy by animal Summary for MTDFRUN, VC and genetic parameters Fixed effect solutions PBV solutions Other random effect solutions Blocks of inverse, F-tests Contrasts, t-tests Expected values of solutions
COMPILING and LINKING FORTRAN code needs to be converted to an executable program in 'machine' language Portable Flexible (match resources to problem) Need a FORTRAN compiler MS Powerstation >>Digital>> Compaq Lahey G77(DOS, LINUX) Usual: *.FOR >> *.OBJ (subroutines, programs) *.FOR >> *.OBJ >>LINK>> *.EXE (subroutines linked to program)
Optimization switches (debugging vs speed) Switches in SETUP.BAT and COMPILE.BAT are for speed not debugging "Include" statement for a file The file is compiled with rest of code PARAM.DAT sets DIMENSIONS of analysis (matrices, etc.) After changes in PARAM.DAT must recompile
MAIN PROGRAMS with SUBROUTINES MTDFNRM.FOR Pedigree file to A-inverse MSTIME.FOR MTDFSUB.FOR (Timer subroutines) (Many subroutines) MTDFPREP.FOR Converts data to standard form (equation no., etc.) Include 'PARAM.DAT' MSTIME.FOR MTDFSUB.FOR
MTDFRUN.FOR Sets up MME, solves, SIMPLEX algorithm for VC Include 'PARAM.DAT' MSTIME.FOR MTDFSUB.FOR SPARS4.FOR SPARS4A.FOR MTDFLIK.FOR (subroutine to calculate -2logL, etc.) Include 'PARAM.DAT' (SPARSPAK subroutines: reorder, Choleski factor and solve) SETUP.BAT COMPILE.BAT Most subroutines to *.OBJ MTDFLIK.FOR to *.OBJ; MTDFNRM.FOR, MTDFPREP.FOR, MTDFRUN.FOR, compiled and linked to MTDFNRM.EXE, MTDFPREP.EXE, MTDFRUN.EXE
c This file is 'param.dat' with code that is included in compiling c mtdfprep.for, mtdflik.for, and mtdfrun.for PARAMETER( c no. traits * MAXTRT=2, c no. of integer variables on record (fixed and random effects), * MAXINTR=30, c no. of real variables on record (covariates and traits) * MAXR8=30, c no. of covariates for each trait * MAXCOV=1, c no. of regression coefficients for each covariate * MAXNFR=1, c no. of fixed effects for each trait * MAXFIX=3, c no. of levels for each fixed effect * MAXNFL=50, c no. of constraints for equations * MAXCONS=1, c no. of animals for each trait * MAXANIM=600, c maximum observations needed for AI matrix * MAXOBS = 2000, c no. of additional uncorrelated random effects for each trait * MAXRAN=3, c no. of levels for each additional uncorrelated random effect * MAXNRL=500, c no. of non-zero elements in coefficient matrix used for c fspak version only + MAXNZE=525000, c length of hash vector used for fspak version only + NHASH=650000, c for sampling variances + MAXINV=40, c length of S vector for SPARSPAK version only * MAXORDS=1250000, c ------------------------ changes to here ----------------------------
rem Reminder that this file is 'setup.bat'; 'rem' is not active command rem Note that '/G4' will be ignored FL32 /c /G4 /Op /D "NDEBUG" spars4a.for FL32 /c /W0 /G4 /Ox /Op /D "NDEBUG" spars4.for FL32 /c /G4 /Ox /Op /D "NDEBUG" mstime.for FL32 /c /G4 /Ox /Op /D "NDEBUG" mtdfsub.for call compile.bat
rem Reminder that this is file 'compile.bat', 'rem' is not active FL32 /G4 /Ox /Op /D "NDEBUG" mtdfnrm.for mtdfsub.obj mstime.obj FL32 /G4 /Ox /Op /D "NDEBUG" mtdfprep.for mtdfsub.obj mstime.obj FL32 /c /G4 /Ox /Op /D "NDEBUG" mtdflik.for FL32 /c /G4 /Ox /Op /D "NDEBUG" mtdfrun.for FL32 /G4 /Ox /Op /D "NDEBUG" mtdfrun.obj mtdflik.obj mstime.obj mtdfsub.obj spars4.obj spars4a.obj rem '/G4' will be ignored
rem The is MOUSE.BAT a batch file that calls answer files for rem a complete run with one restart of MTDFREML rem rem is a memo for DOS rem whatever is to the right of < is used as an answer file rem in place of answers typed on keyboard mtdfnrm<mousenrm.in rem copy to save mtdf56 to mtdf56.1 (on next run old mtdf56 is replaced) copy mtdf56 mtdf56.1 rem < answer file for mtdfprep--can be any name mtdfprep<mouseprp.in rem copy to save mtdf66 to mtdf66.1 which is log file for mtdfprep copy mtdf66 mtdf66.1
rem < answer file for initial start for mtdfrun--can be any name mtdfrun<mouserun.in rem copy mtdf4 to mtdf4.1 for restart of mtdfrun with mtdf4.1 as answer copy mtdf4 mtdf4.1 rem copy to save mtdf76 to mtdf76.1 to check -2log L on next restart copy mtdf76 mtdf76.1 rem < answer file from previous start updated with new estimates mtdfrun<mtdf4.1 copy mtdf4 mtdf4.2 rem copy to save to check -2log L and estimates for global converge copy mtdf76 mtdf76.2 rem < answer file to use previous estimates to solve MME and rem to calculate SE of contrasts and expectations of solutions mtdfrun<mouserun.se rem copy mtdf67 to mtdf67.1 to save file of contrasts and expectations copy mtdf67 mtdf67.1 rem copy mtdf77 to mtdf77.1 to save fixed effects solutions copy mtdf77 mtdf77.1
20101 11012 10101 0 11012 615 701 0 10101 0 0 0 20102 11012 10101 0 20103 11012 10101 0 20104 11012 10101 0 20112 11012 10101 0 20113 11012 10101 0 20114 11012 10101 0 20115 11012 10101 0 20302 10614 10506 0 10614 2315 2201 0 10506 1813 1701 0 20303 10614 10506 0 20304 10614 10506 0 20306 10614 10506 0 20312 10614 10506 0 20313 10614 10506 0 20314 10614 10506 0 20401 10813 10701 0 . . . . . . . . . . . . . . This is 'mouse.ped' Animal Sire Dam Zero (not needed) Does not need to be ordered Can have duplicates An ID > Sire and Dam ID Zero ID is unknown For sire model would have ped file SireID 0 0 Can use field 2, and 4 twice
41601 31514 31602 0 41602 31514 31602 0 41603 31514 31602 0 41604 31514 31602 0 41612 31514 31602 0 41613 31514 31602 0 41614 31514 31602 0 41615 31514 31602 0
0 0 for animal model,1 for sire/MGS model [This is 'mousenrm.in'] 41615 maximum id number represented in pedigree file, can be larger 1 minimum id number represented in pedigree file, can be 0 mouse.ped name of pedigree file 1 write inbreeding file to mtdf13, 0 would not write that file 4 no. integer fields in ped file (must be no reals before them)* 1 position of animal (or sire for sire, sire of sire, mgs of sire) 2 position of sire (or sire of sire) 3 position of dam (or MGS of sire) 0 number of genetic (Westell) groups
Started 07:05:12.17 on 08/10/1998 FILE is MTDF56 from MTDFNRM +++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++ PROGRAM "MTDFNRM" - Calculate A-1 for "MTFRUN" and recode IDs for "MTDFPREP" Version to use Westell grouping strategy +++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++ OPTION FOR CALCULATION OF A-1 FOR ANIMAL SIRE DAM TYPE .... 0 FOR ANIMAL SIRE MGS TYPE ... 1 OPTION CHOSEN FOR THIS ANALYSIS = 0 MAXIMUM ID = 41615 MINIMUM ID = 1 PEDIGREE FILE OPENED, IUN33 = mouse.ped ************ REORDERED ANIMAL FILE OPENED, IUN11 = MTDF11 FILE FOR A-1 ELEMENTS OPENED, IUN44 = MTDF44 FILE FOR IDS AND INBREEDING COEFFICIENTS OPENED THIS FILE WILL CONTAIN ANIMAL, SIRE, AND DAM RECODED AND ORIGINAL IDS FOLLOWED BY THE INBREEDING COEFFICIENT FOR EACH NO. INTEGER FIELDS PER RECORD IN IUN33 = 4 ANIMAL ID IN POSITION ...... 1 SIRE ID IN POSITION ........ 2 DAM (MGS) ID IN POSITION ... 3 NO. OF GENETIC GROUPS FOR CALCULATION OF W = 0 The current time is: 07:05:12.72 NO. OF PEDIGREES READ = 309 NO. OF DIFFERENT ANIMALS = 329 INCLUDES NO. OF GENETIC GROUPS = 0
END OF FIRST PASS The current time is: 07:05:12.89 END OF SORT The current time is: 07:05:12.89 FIRST 10 REORDERED IDs 1 215 FIRST 10 REORDERED IDs 2 403 FIRST 10 REORDERED IDs 3 615 FIRST 10 REORDERED IDs 4 701 ID VECTOR WRITTEN IN ORDER TO IUN11 The current time is: 07:05:13.05 SIRE AND DAM IN PEDIGREE REORDERED IN IVECS AND IVECD The current time is: 07:05:13.33 CALCULATION OF A-1 FROM ANIMAL SIRE DAM (IOPT = 0) NON-ZERO HS ELEMENTS FOR NRM INVERSE = 1241 LOG DETERMINANT OF NRM = -210.71674289 NUMBER OF INBRED ANIMALS = 0 ... WITH AVERAGE INBREEDING COEF = .00000000 TOTAL NO. OF ANIMALS INCLUDING BASE AND GENETIC GROUPS = 329 ******* The current time is: 07:05:13.99 The elapsed time was: 00:00:01.27
329 (No.) [ MTDF13 optional for an, sire, dam codes and F] RECODED ORIGINAL F = inbreeding AN SIRE DAM AN SIRE DAM AN SIRE DAM 1 0 0 215 0 0 .000 .000 .000 2 0 0 403 0 0 .000 .000 .000 3 0 0 615 0 0 .000 .000 .000 4 0 0 701 0 0 .000 .000 .000 5 0 0 814 0 0 .000 .000 .000 6 0 0 904 0 0 .000 .000 .000 7 0 0 1314 0 0 .000 .000 .000 8 0 0 1602 0 0 .000 .000 .000 9 0 0 1701 0 0 .000 .000 .000 10 0 0 1813 0 0 .000 .000 .000 11 0 0 2201 0 0 .000 .000 .000 12 0 0 2315 0 0 .000 .000 .000 13 0 0 2401 0 0 .000 .000 .000
23 0 0 10404 0 0 .000 .000 .000 24 10 9 10506 1813 1701 .000 .000 .000 25 10 9 10512 1813 1701 .000 .000 .000 26 12 11 10602 2315 2201 .000 .000 .000 27 12 11 10614 2315 2201 .000 .000 .000 28 16 13 10701 2512 2401 .000 .000 .000 29 16 13 10715 2512 2401 .000 .000 .000 30 20 17 10801 3015 2904 .000 .000 .000 31 20 17 10813 3015 2904 .000 .000 .000 32 1 2 10902 215 403 .000 .000 .000 33 1 2 10917 215 403 .000 .000 .000 34 3 4 11002 615 701 .000 .000 .000 40 0 0 11316 0 0 .000 .000 .000 41 0 0 11404 0 0 .000 .000 .000 42 14 15 11505 2416 2503 .000 .000 .000 . . . 329 214 216 41615 31514 31602 .000 .000 .000
This is 'mouse.dat' Animal Sire Dam Gen Sex LS Lit LS* T-1 T-2 _____________________________________________________ 20101 11012 10101 1 1 4 1 4.0 22.5 59.1 20102 11012 10101 1 1 4 1 4.0 22.6 52.4 20103 11012 10101 1 1 4 1 4.0 22.9 61.1 20104 11012 10101 1 1 4 1 4.0 23.0 57.9 20112 11012 10101 1 2 4 1 4.0 24.6 69.3 20113 11012 10101 1 2 4 1 4.0 26.4 66.4 20114 11012 10101 1 2 4 1 4.0 24.1 61.6 20115 11012 10101 1 2 4 1 4.0 24.3 68.3 20302 10614 10506 1 1 7 2 7.0 23.6 60.1 20303 10614 10506 1 1 7 2 7.0 24.2 60.8 20304 10614 10506 1 1 7 2 7.0 22.5 62.8 20306 10614 10506 1 1 7 2 7.0 22.3 59.4 20312 10614 10506 1 2 7 2 7.0 23.7 66.9 20313 10614 10506 1 2 7 2 7.0 27.5 72.9 20314 10614 10506 1 2 7 2 7.0 17.3 58.4 20401 10813 10701 1 1 2 3 2.0 20.3 58.1
20402 10813 10701 1 1 2 3 2.0 21.3 58.7 20403 10813 10701 1 1 2 3 2.0 21.9 53.2 20404 10813 10701 1 1 2 3 2.0 22.1 57.1 20413 10813 10701 1 2 2 3 2.0 28.8 62.5 20501 11012 10902 1 1 7 4 7.0 18.9 59.3 20503 11012 10902 1 1 7 4 7.0 23.2 69.0 . . . . . . . . . . . . . . 41604 31514 31602 3 1 3 42 3.0 25.1 62.8 41612 31514 31602 3 2 3 42 3.0 23.3 66.6 41613 31514 31602 3 2 3 42 3.0 26.0 60.6 41614 31514 31602 3 2 3 42 3.0 27.0 65.2 41615 31514 31602 3 2 3 42 3.0 26.3 60.7
mouse.dat name of data file Mouse data from Karin Meyer [first of comment lines] [This is mouseprp.in] mouse.dat [will now show file name in mtdf66] * end of comments 7 number 'integer' fields 3 number 'real' fields 1 number of traits to analyze body weight name of first trait 2 position of trait in vector of 'reals' 0. missing value indicator, cannot be '.' 0 number of covariates 3 number of fixed factors generation name of first fixed factor 4 position in vector of 'integers' 1 *** write summary of levels of factor, O would not write sex name of second fixed factor 5 position in vector of 'integers' 1 write summary
litter size name of third fixed factor 6 position in vector of 'integers' 1 write summary 1 position of animal ID in vector of 'integers' 329 *** number of animals (in pedigree from mtdfnrm, mtdf11, mtdf56 1 second animal effect (usually maternal genetic of dam) maternal genetic effect name for second animal effect 3 position of second animal effect in vector of 'integers' 1 number of other random effects maternal permanent environment name of random effect 3 position of random effect in vector of 'integers' 0 *** don't write out summary, usually a big file 1 *** write out file to match fixed factor levels with solutions 1 *** write out file to match other random levels with solutions
MTDF66 from MTDFPREP Started 07:05:16.51 on 08/10/1998 PROGRAM "MTDFPREP" - Setup W=X:Z matrix for MT-IAM Data set description : Mouse data from Karin Meyer single trait analysis mouse.ped No. of data lines in Unit 33 = 284 No. of integer variables per record = 7 No. of real variables per record = 3 No. of traits = 1 No. of valid records = 284 No. of animals in A-1 = 329 Order of MME (before constraints) = 712 -------------------------------------------------------------------------------- Continued
Results for trait 1 - body weight (position 2 ) No. of records = 284 (missing value: .0000 No. missing = 0 ) Trait Mean SD CV Min Max Std Min Std Max 1 24.0687 3.30236 13.72 14.600 34.500 -2.87 3.16 No. of covariates = 0 No. of fixed effects = 3 1: 3 levels for generation (MME rows: 1 - 3) Level Value No. % Mean 1 1 93 32.75 23.724 2 2 84 29.58 23.063 3 3 107 37.68 25.158 2: 2 levels for sex (MME rows: 4 - 5) Level Value No. % Mean 1 1 150 52.82 22.656 2 2 134 47.18 25.650 3: 7 levels for litter size (MME rows: 6 - 12) Level Value No. % Mean 1 1 11 3.87 26.609 2 2 41 14.44 23.722 7 7 30 10.56 21.973 No. of animals in A-1 = 329 (MME rows: 13 - 341) No. of 2nd animal effects = 1 1: 329 levels for maternal genetic eff (MME rows: 342 - 670) No. of uncorrelated random effects = 1 1: 42 levels for maternal perm env (MME rows: 671 - 712) -------------------------------------------------------------------------------- Continued
MTDF66 from MTDFPREP Continued Order of MME (before constraints) = 712 Fixed effects = 3 Trait No. Name Position Levels Rows 1 1 generation 4 3 1 - 3 1 2 sex 5 2 4 - 5 1 3 litter size 6 7 6 - 12 Animal effects = 1 Trait No. Name Position Levels Rows 1 1 Animal w/ full A-1 1 329 13 - 341 2nd Animal effects = 1 Trait No. Name Position Levels Rows 1 1 maternal genetic eff 3 329 342 - 670 Uncorrelated random effects = 1 Trait No. Name Position Levels Rows 1 1 maternal perm env 3 42 671 - 712 -------------------------------------------------------------------------------- Files written:
Mouse data from Karin Meyer {This file is 'mouserun.in'] single trait analysis * end of comments 0 *** for start or restart, 1 for continue or solns from previous run 1 option to estimate variance components, for solns to MME use 4 0 number of constraints to be imposed by you 0 *** not reordered previously, if reordered can use 1 if mtdf58 saved 1 3.65354 starting value for a1,a1 2 .5359 starting value for m1,a1*** 3 1.37502 starting value for m1,m1 0 0 end input for G 1 yes, values on screen are correct 0 number to be held constant 1 .426651 starting value for c1,c1 other random factor 0 0 end input for C 1 yes, values on screen are correct 0 number to be held constant 1 2.28714 starting value for e1,e1 residual variance 0 0 end input for R 1 yes, values on screen are correct 0 number to be held constant 1 *** write solutions for covariates and fixed factors 1 *** match original codes of fixed factor levels with solns 1 *** write solutions for animal effects 1 *** write solutions for other random factors 1 *** match original codes of random factor levels 1.d-6 convergence for variance of simplex -2logL 300 number of simplex rounds
MTDF76 from MTDFRUN Started 07:05:23.98 on 08/10/1998 ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++ PROGRAM "MTDFRUN" - Estimate Covariance Components for MT-IAM ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++ Mouse data from Karin Meyer single trait analysis Cold start, i.e., not a continuation of previous run Run option 1: iterate for variance components 0 constraints imposed by user ****** (Co)variances in model: No. in likelihood calculation = 5 No. to be held constant = 0 No. to be maximized = 5 Starting values for this run: G matrix: 4.0 .5 .5 1.5 C matrix: .5 R matrix: 2.0 ** reordering called ** ** reordering completed ** The elapsed time was: 00:00:00.39 ** solve5 called ** ** solve5 completed ** The elapsed time was: 00:00:00.99 Continue............
SPARSPAK-A statistics.. Time: Ordering = .270 secs. ( .004 mins.) Total/Solution = .170 secs. ( .003 mins.) Allocation = .000 secs. ( .000 mins.) Factorization = .109 secs. ( .002 mins.) Solve = .061 secs. ( .001 mins.) Storage: No. equations = 712 Non-zero hs elements in MME = 9742 Maximum storage required = 12304. ( .094 MB) Size of storage array (MAXSA) = 800000 ( 1.538% used) ******* RESULTS FROM SIMPLEX ******* OPTIONS SET FOR THIS RUN: MAXIMUM NO. OF SIMPLEX ITERATES ALLOWED = 300 MINIMUM VARIANCE OF FUNCTION VALUES IN SIMPLEX = .1000000000E-05 NO. OF SIMPLEX ITERATIONS CARRIED OUT = 44 NO. OF LIKELIHOODS EVALUATED = 79 NO. OF NON-PERMISSABLE PARAMETER VECTORS = 0 No. of failed contractions = 0 Variance of simplex function values = .2190791759E-06 Convergence criterion attained Continue............
MTDF76 continued Final Simplex: (++ best L; *** parameter held constant) 1 753.7488165 3.6828 .5114 1.3544 .4240 2.2586 2 753.7498457 3.6449 .4895 1.4246 .3877 2.2988 ++ 5 753.7486078 3.6407 .4745 1.3861 .4140 2.2980 6 753.7494189 3.7354 .5228 1.3221 .4393 2.2565 -2 log L = 753.7486077658 ( 5) Var = .0000002191 Half-store elements of AI inverse [Sample variances of estimates] 1 1 7.953967229489519 1 2 -3.619170958644206 1 5 -3.902998228002057 2 2 3.884627165149515 2 3 -3.275798374041433 2 5 1.821341783889839 3 3 6.128186867862764 3 4 -3.214987941640724 3 5 -1.224458463913176 4 4 2.092185282214288 4 5 5.792220185962393E-001 5 5 2.054178793146739
0 Constraints: Estimates: GENETIC VARIANCES AND COVARIANCES : a1 m1 a1 : 3.64066 .47452 m1 : .47452 1.38608 UNCORRELATED RANDOM VARIANCES AND COVARIANCES: T1 C3 T1 C3 : .414033 ENVIRONMENTAL VARIANCES AND COVARIANCES : e1 e1 : 2.29804 PHENOTYPIC VARIANCES AND COVARIANCES : p1 p1 : 8.21333 HERITABILITIES AND GENETIC CORRELATIONS a1 m1 a1 : .44 ( .320) m1 : .21 .17 ( .712) ( .296) UNCORRELATED RANDOM EFFECTS T1 C3 T1 C3 : .50E-01 ( .177)
MTDF76 continued ENVIRONMENTAL PROPORTION OF TOTAL VARIANCE AND CORRELATIONS e1 e1 : .28 ( .196) Files written: MTDF4 (ascii): Parameter file (IUN5) for "cold" restart MTDF54 (ascii): Last simplex MTDF58 (binary): SPARSPAK reordering MTDF59 (ascii): Constraints imposed MTDF68 (ascii): Likelihoods by rounds MTDF67 (ascii): Sampling variances if requested MTDF72 (ascii): Predicted BVs and PEVs if requested MTDF76 (ascii): Program log file MTDF77 (ascii): Solutions for covariates and fixed effects if requested MTDF78 (ascii): Solutions for trait within animal if requested MTDF79 (ascii): Solutions for independent random effects if requested The current time is: 07:06:25.61 The elapsed time was: 00:01:00.64
Mouse data from K Meyer (This was mtdf4 copied to mtdf4.1 for restart) single trait analysis * end of comments 0 continuation: 0-no; 1-yes 1 run option 0 # constraints 0 reordered: 0-no; 1-yes <<< could change to 1 1 3.637814264727246 animal effect prior 2 5.289616129513786E-001 animal effect prior 3 1.300806074847860 animal effect prior 0 0.D0 1 values are correct 0 # parameters to hold constant 1 4.647326260772600E-001 uncorrelated effect prior 0 0.D0 1 values are correct 0 # parameters to hold constant 1 2.290805075973958 residual effect prior 0 0.D0 1 values are correct 0 # parameters to hold constant 1 write fixed effect solutions 1 merge PREP info with solutions 1 write animal solutions?? 1 write independent random effect solutions 1 merge PREP info with solutions 1.000000000000000E-006 convergence criterion <<< could change 300 # Simplex rounds <<< could change
MTDF76.2 after restart to check change in -2logL Started 14:10:31.49 on 08/26/2000 ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++ PROGRAM "MTDFRUN" - Estimate Covariance Components for MT-IAM Mouse data from Karin Meyer single trait analysis Cold start, i.e., not a continuation of previous run Run option 1: iterate for variance components Starting values for this run: G matrix: 3.6 .5 .5 1.3 C matrix: .5 R matrix: 2.3 ** reordering called and completed** MAXIMUM NO. OF SIMPLEX ITERATES ALLOWED = 300 MINIMUM VARIANCE OF FUNCTION VALUES IN SIMPLEX = .1000000000E-05 NO. OF SIMPLEX ITERATIONS CARRIED OUT = 15 NO. OF LIKELIHOODS EVALUATED = 34 Variance of simplex function values = .7619503856E-06 Convergence criterion attained Final Simplex: (++ best L; *** parameter held constant) ++ 1 753.7462824 3.6378 .5290 1.3008 .4647 2.2908 6 753.7472056 3.6534 .5759 1.2561 .4875 2.2994 -2 log L = >>> 753.7462824159 <<< (1) Var = .0000007620 Large Sample Variances of Variance Estimates Continued
MTDF76.2 continued Estimates: GENETIC VARIANCES AND COVARIANCES : a1 m1 a1 : 3.63781 .52896 m1 : .52896 1.30081 UNCORRELATED RANDOM VARIANCES AND COVARIANCES: T1 C3 T1 C3 : .464733 ENVIRONMENTAL VARIANCES AND COVARIANCES : e1 e1 : 2.29081 PHENOTYPIC VARIANCES AND COVARIANCES : p1 p1 : 8.22312 HERITABILITIES AND GENETIC CORRELATIONS a1 m1 a1 : .44 ( .320) m1 : .24 .16 (1.137) ( .296) UNCORRELATED RANDOM EFFECTS T1 C3 T1 C3 : .57E-01 ( .178) ENVIRONMENTAL PROPORTION OF TOTAL VARIANCE AND CORRELATIONS e1 e1 : .28 ( .196) Files written: The elapsed time was: 00:00:01.54
MTDF77 [Fixed effect solutions] ______________________________________________________ solution level * name trait 1 -.16211010 1 * generation * body weight 2 -1.64614075 2 * generation * body weight 3 .01080553 3 * generation * body weight 4 -3.05158974 1 * sex * body weight 5 .00000000 <<<< 2 * sex * body weight 6 5.73575613 1 * litter size * body weight 7 2.22139506 2 * litter size * body weight 8 3.28421069 3 * litter size * body weight 9 1.34509621 4 * litter size * body weight 10 2.27238624 5 * litter size * body weight 11 1.53586287 6 * litter size * body weight 12 .00000000 <<<< 7 * litter size * body weight
MTDF79 [Uncorrelated random effect solutions] equn solution level * name trait _________________________________________________________ 671 -.04583612 10101 * maternal permanent e * body weight 672 .07454918 10404 * maternal permanent e * body weight 673 -.03016894 10506 * maternal permanent e * body weight 674 .03943667 10602 * maternal permanent e * body weight 675 -.08974688 10701 * maternal permanent e * body weight 676 -.09208224 10801 * maternal permanent e * body weight 704 -.09045377 30902 * maternal permanent e * body weight 705 -.18210848 30904 * maternal permanent e * body weight 706 -.00342698 31105 * maternal permanent e * body weight 707 .02015426 31203 * maternal permanent e * body weight 708 -.24623957 31302 * maternal permanent e * body weight 709 -.19488994 31304 * maternal permanent e * body weight 710 .13085314 31401 * maternal permanent e * body weight 711 .26078214 31501 * maternal permanent e * body weight 712 -.12766108 31602 * maternal permanent e * body weight
MTDF78 [Genetic solutions] Coded ID ID a1 m1 ___________________________________________________ 1 215 .77065064 .29387514 2 403 .77065064 .29387514 3 615 1.03862464 .49508782 4 701 1.03862464 .49508782 5 814 -.42773805 -.23195017 6 904 -.42773805 -.23195017 7 1314 -.39957873 -.16270164 8 1602 -.39957873 -.16270164 9 1701 .46445741 .46909996 10 1813 .46445741 .46909996 11 2201 .50407019 .56651447 12 2315 .50407019 .56651447 13 2401 -.12542643 -.11625806 14 2416 -1.52961347 -1.11182385 . . . . . . . . Continued
MTDF78 [Genetic solutions] continued 318 41512 .98956176 .40385134 319 41513 .28142180 .30088323 320 41516 2.22880667 .58404553 321 41517 1.96325419 .54543249 322 41601 .20518050 -.31031653 323 41602 1.04609669 -.18804190 324 41603 .24943925 -.30388102 325 41604 .86906171 -.21378393 326 41612 -1.27819113 -.52600831 327 41613 -.08320496 -.35224963 328 41614 .35938251 -.28789456 329 41615 .04957128 -.33294311 [For all animals in pedigree file ]
This answer file is 'mouserun.se' to do option 4 Note uses continuation option to read converged variance components Calculate F-stats, 'naive' t-stats for contrasts, and expectations * 1 continuation: 0-no; 1-yes [Files must be available] 4 run option 1 write fixed solutions 1 match with prep ID 0 do not write animal solutions 0 do not write other random factor solutions 1 calculate block of inverse and F-statistic 4 first equation of block for sex 5 last equation of block for sex 1 calculate block of inverse for litter size 6 first equation for block for litter size 12 last equation for block for litter size 0 do not calculate another block of inverse of coef matrix
1 calculate a contrast for sex 1 equn 4 vs sex 2 equn 5 2 number of elements in contrast 4 1. equn and coef for sex 1 t stat squared = F 5 -1. equn and coef for sex 2 1 calculate contrast for generation 1 vs 2 2 number of elements in contrast 1 1. equn and coef for generation 1 2 -1. equn and coef for generation 2 1 calculate contrast for generations 1 and 2 vs 3 3 number of elements in contrast 1 1. equn and coef for generation 1 2 1. equn and coef for generation 2 3 -2. equn and coef for generation 3 0 no more contrasts
0 no PEV and rTI 1 calculate expectations of solutions 1 equation corresponding to first parameter in expectation 12 " " last " " 1 equation corresponding to first solution in block for gen and sex 5 " " last " " 1 calculate more expectations 1 first parameter in expectation 12 last parameter in expectation 6 first solution in block for litter size 12 last solution in block 0 no more expectations
MTDF67 from option 4 F-tests, t-tests, expectations Block of inverse for equations 4 to 5 4 .0608 .0000 5 .0000 .0000 inverse of inverse of order 2 with rank 1 F=statistic = 153.186 for eqn 4 to 5 with n = 2 but DF = 1 Block of inverse for equations 6 to 12 6 1.8511 .6086 .6306 .4512 .6347 .7711 .0000 7 .6086 1.2548 .6558 .8239 .8051 .7763 .0000 8 .6306 .6558 1.4760 .4720 .6430 .7432 .0000 9 .4512 .8239 .4720 1.4383 .7375 .6339 .0000 10 .6347 .8051 .6430 .7375 1.0013 .7082 .0000 11 .7711 .7763 .7432 .6339 .7082 1.3437 .0000 12 .0000 .0000 .0000 .0000 .0000 .0000 .0000 inverse of inverse of order 7 with rank 6 F=statistic = 3.617 for eqn 6 to 12 with n = 7 but DF = 6 Continued
Contrast 1 with no. elements 2 Row 4 coefficient 1.000000000000000 Row 5 coefficient -1.000000000000000 Contrast 1 =-.30515897E+01 SE = .24655668E+00 and t = -12.38 Contrast 2 with no. elements 2 Row 1 coefficient 1.000000000000000 Row 2 coefficient -1.000000000000000 Contrast 2 = .14840307E+01 SE = .64213778E+00 and t = 2.31 Contrast 3 with no. elements 3 Row 1 coefficient 1.000000000000000 Row 2 coefficient 1.000000000000000 Row 3 coefficient -2.000000000000000 Contrast 3 =-.18298619E+01 SE = .11472325E+01 and t = -1.60 Continued
MTDF67 from option 4 F-tests, t-tests, expectations continued Contributions of parameters 1 to 12 To expectations for solutions 1 to 5 Parameter: Coefficients for solutions 1 to 5 1 1.000 .000 .000 .000 .000 2 .000 1.000 .000 .000 .000 3 .000 .000 1.000 .000 .000 4 .000 .000 .000 1.000 .000 5 1.000 1.000 1.000 -1.000 .000 6 .000 .000 .000 .000 .000 10 .000 .000 .000 .000 .000 11 .000 .000 .000 .000 .000 12 1.000 1.000 1.000 .000 .000
Contributions of parameters 1 to 12 To expectations for solutions 6 to 12 Parameter: Coefficients for solutions 6 to 12 1 .000 .000 .000 .000 .000 .000 .000 5 .000 .000 .000 .000 .000 .000 .000 6 1.000 .000 .000 .000 .000 .000 .000 7 .000 1.000 .000 .000 .000 .000 .000 8 .000 .000 1.000 .000 .000 .000 .000 9 .000 .000 .000 1.000 .000 .000 .000 10 .000 .000 .000 .000 1.000 .000 .000 11 .000 .000 .000 .000 .000 1.000 .000 12 -1.000 -1.000 -1.000 -1.000 -1.000 -1.000 .000
mouse.dat Mouse data from Karin Meyer (This file is 'mouseprp.in2' two trait analysis * end of comments 7 number 'integer' fields 3 number 'real' fields 2 number of traits to analyze body weight name of first trait 2 position of trait in vector of 'reals' 0. missing value indicator, cannot be '.' 0 number of covariates 3 number of fixed factors generation name of first fixed factor 4 position in vector of 'integers' 1 write summary of levels of factor, O would not write sex name of second fixed factor 5 position in vector of 'integers' 1 write summary litter size name of third fixed factor 6 position in vector of 'integers' Continued
litter size name of third fixed factor 6 position in vector of 'integers' 1 write summary 1 postion of animal ID in vector of 'integers' 329 number of animals (in pedigree from mtdfnrm, mtdf11, mtdf56 1 second animal effect (usually maternal genetic of dam) maternal genetic effect name for second animal effect 3 position of second animal effect in vector of 'integers' 1 number of other random effects maternal permanent environment name of random effect 3 position of random effect in vector of 'integers' 0 don't write out summary, usually a big file Continued
mouse.dat Mouse data from Karin Meyer (This file is 'mouseprp.in2' two trait analysis Continued) other trait 3 position 0. missing value indicator 0 covariates 3 fixed factors generation 4 position 1 write summary sex 5 position 1 write summary lit size 6 position 1 write summary 1 second animal effect mat gen 2 (name) 3 position 1 other random factor mat pe (name) 3 position 0 do not write summary 1 write file to match fixed factor levels with solutions 1 write file to match other random levels with solutions
Mouse data from Karin Meyer (This file is 'mouserun.in2') two trait analysis * end of comments 0 for start or restart, 1 for continue or solns from previous run 1 option to estimate variance components, for solns to MME use 4 0 number of constraints to be imposed by you 0 not reordered previously,if reordered can use 1 if mtdf58 saved 1 4. starting value for a1,a1 2 3.5 a1,a2 3 .5 starting value for m1,a1 4 .4 a1,m2 5 20. a2,a2 6 .1 a2,m1 7 2. a2,m2 8 1.5 starting value for m1,m1 9 .1 a1,m2 10 7. m2,m2 0 0 end input for G 1 yes, values on screen are correct 0 number to be held constant