Juice Weight of Sugarcane in Pest Conditions
Explore the impact of 11 pest conditions on the juice weight of sugarcane through a one-way ANOVA analysis. Gain insights into the statistical effect of pests on cane yield and juice quality based on a study by Mahalanobis and Bose. Understand the experimental setup, treatments, and responses to assess the variations. Discover the estimated weighted least squares and basic calculations for F-test and WLS F-stat.
Uploaded on | 0 Views
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
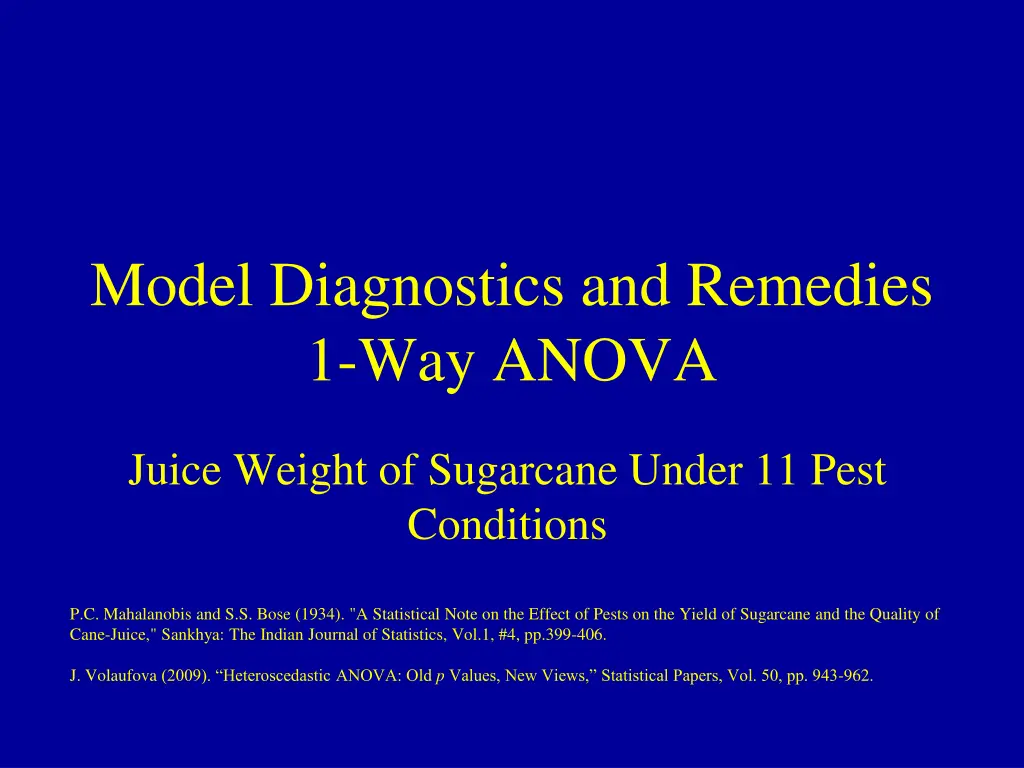
Model Diagnostics and Remedies 1-Way ANOVA Juice Weight of Sugarcane Under 11 Pest Conditions P.C. Mahalanobis and S.S. Bose (1934). "A Statistical Note on the Effect of Pests on the Yield of Sugarcane and the Quality of Cane-Juice," Sankhya: The Indian Journal of Statistics, Vol.1, #4, pp.399-406. J. Volaufova (2009). Heteroscedastic ANOVA: Old p Values, New Views, Statistical Papers, Vol. 50, pp. 943-962.
Data Description Experimental units=25 grouped canes, 6 reps per treatment Responses: Weight of Canes, Weight of Juice (lbs) Treatments: 1=Healthy (Control) 2=Top-shoot Borer 3=Stem Borer 4=Top-shoot & Stem Borer 5=Root Borer 6=Termites 7=Stem & Root Borer 8=Root Borer & Termites 9=Top-Shoot & Root Borer 10=Top-Shoot & Stem & Root Borer 11=Top-Shoot Borer & Termites
Introduction Population Variance: Measure of average squared deviation of individual measurements around the mean N 2 ( ) Y i ( ) ( ) f y dy 2 = = = = 2 2 = ( ) [( ) ] 1 i V Y E Y y N Sample Variance: Measure of average squared deviation of a sample of measurements around their sample mean. Unbiased estimator of 2 ( 2 1 i S n ) n 2 Y Y i = = 1
Estimated Weighted Least Squares 0 Y 1 i ( ) Y Y = + = = = = = 2 i 2 i Y 1 I 1,..., ; 1,..., ~ 0, independent Y i g j n N E V ij i ij i ij i i i n i n i i Y in i 2 1 I 0 0 1 0 1 n n I n n n n n n n 1 1 1 2 1 g 1 1 1 1 Y 0 1 0 1 2 2 1 0 0 Y n n n n 2 n n n n n = = = = = = = Y V X V X 2 2 2 2 2 1 2 2 g Y 0 0 Y g 0 0 1 1 2 g 0 0 I g n n n n n n n n n g g g g 1 2 g g g ( ) ( ) 1 1 1/2 Y 1/2 Y 1/2 Y = = = = = * * * 0 * * * * * * 0 * 0 * 0 * 0 * 0 Y V Y X V X X V X P X X X X P X X X X Define: ' ' ' ' 0 W SS g SS N Trt 1 H ( ) ( ) 0 = = = = = = * * * 0 * * * * W W Y P P Y Y I P Y To test : ... : ' ' ~ H SS SS F F N g 0 1 Tr t Err 1, g W g W Err g 2 1 I 0 0 s EW SS g SS N n n n n n 1 1 I 2 1 g Trt 2 2 0 0 s 1 ^ n n n n n = = V V Estimated Weighted Least Squares: Replace with F 2 1 2 2 g Y Y EW EW Err g 2 g 0 0 I s n n n n n 1 2 g g g
Basic Calculations F-Test and WLS F-Stat trt n.trt ybar.trt 6 42.91667 0.641667 6 28.88333 1.805667 8.230384 9.028334 3.322872 2772.095 95.97562 6 29.73333 4.450667 24.51172 22.25333 1.348113 1191.827 40.08388 6 17.35 6.787 644.2413 33.935 0.884043 266.1168 15.33815 6 27.56667 13.55067 0.126936 67.75333 0.442783 6 30.25 2.975 38.64498 14.875 2.016807 1845.504 6 26.81667 2.305667 4.810994 11.52833 2.602284 6 29.48333 8.145667 18.82309 40.72833 0.736588 640.2914 21.71707 6 22.08333 11.70167 190.0997 58.50833 0.512747 250.0533 11.32317 6 25.71667 15.44567 23.89095 77.22833 0.388458 256.9059 9.989858 6 24.03333 20.10667 81.20102 100.5333 0.298408 66 2421.651 439.58 21.90375 26825.43 745.8972 27.71212 7.992364 s2.trt SS_TRT 1387.07 3.208334 9.350649 SS_ERR w=n/s2 w*ybar^2 w*ybar 17222.4 401.2987 1 2 3 4 5 6 7 8 9 336.48 12.20604 61.0084 69.7846 1871.39 10 11 172.361 7.17175 Sum Mean ANOVA Source Trts Error Total g=11 Trts df SS MS F F(.95) P(>F) 10 2421.6510 242.1651 55 439.5800 65 2861.2310 30.2996 2.0078 0.0000 7.9924 F_EWLS (1/(11-1))*(H13-I13^2/G13) 142.5093
Tests Among g 2 Population Variances Hartley s Fmax Test Must have equal sample sizes (n1= = ng) Test based on assumption of normally distributed data Uses special table for critical values Levene s Test No assumptions regarding sample sizes/distributions Uses F-distribution for the test Bartlett s Test Can be used in general situations with grouped data Test based on assumption of normally distributed data Uses Chi-square distribution for the test
Hartleys Fmax Test H0: 12= = g2 (homogeneous variances) Ha: Population Variances are not all equal Data: smax2 is largest sample variance, smin2 is smallest Test Statistic: Fmax = smax2/smin2 Rejection Region: Fmax F* (Values from class website, indexed by (.05, .01), g (number of populations) and df2 (n-1, where n is the individual sample sizes) Pest Data: Fmax = smax2/smin2 = 20.10667/0.641667=31.34 F*(.05,11,6-1=5) = 28.2 Reject H0
Levenes Test H0: 12= = g2 (homogeneous variances) Ha: Population Variances are not all equal Data: For each group, obtain the following quantities: th the measurement from group ij y j = = ( 1,..., 1,..., ) i i g j n i ~ y = sample median for group ( 1,..., ) i i g i ~ y = = = ( 1,..., 1,..., ) z y i g j n ij ij i i n n g i i z z ij ij = = = 1 n 1 1 j i j = = = + + ... z z N n n . .. i 1 g N i g ( ( ) ) 2 ( ) 1 n z z g . .. i i = = Test Statistic: 1 i L n g 2 i ( ) z z N g . i ij = = 1 1 i L j Rejection Region: F N g , 1, g
Levenes Test Pest data pest <- read.table("http://users.stat.ufl.edu/~winner/data/pests_cane.dat", header=F,col.names=c("pest.trt","wt.cane","wt.juice")); attach(pest) g <- length(unique(pest.trt)) N <- length(pest.trt) mean.trt <- as.vector(tapply(wt.juice, pest.trt, mean)) n.trt <- as.vector(tapply(wt.juice, pest.trt, length)) median.trt <- as.vector(tapply(wt.juice, pest.trt, median)) median.trt.long <- rep(median.trt, n.trt) z <- abs(wt.juice - median.trt.long) mean.trt.z <- as.vector(tapply(z, pest.trt, mean)) mean.trt.z.long <- rep(mean.trt.z, n.trt) mean.all.z <- mean(z) SS.Levene.1 <- sum(n.trt * (mean.trt.z - mean.all.z)^2) SS.Levene.2 <- sum((z-mean.trt.z.long)^2) F.Levene <- (SS.Levene.1 / (g-1)) / (SS.Levene.2 / (N-g)) p.F.Levene <- 1 - pf(F.Levene, g-1, N-g) > Levene.out <- cbind(SS.Levene.1, SS.Levene.2, g-1, N-g, F.Levene, qf(.95,g- 1,N-g), p.F.Levene) > colnames(Levene.out) <- c("SSB", "SSW", "dfB", "dfW", "F", "F(.95)","P(>F)") > round(Levene.out, 4) SSB SSW dfB dfW F F(.95) P(>F) [1,] 44.5115 212.2083 10 55 1.1536 2.0078 0.3414
Bartletts Test General Test that can be used in many settings with groups H0: 12= = g2 (homogeneous variances) Ha: Population Variances are not all equal MSE Pooled Variance g 1 C ) ( ) 1 ln ( ) ( ln ) ( = 2 B 2 i : TS X N g MS n s ERR i = 1 i g 1 1 1 = + 1 C ( ) 3 1 1 g n N g = 1 i i ( ) 2 B 2 2 g 2 B : P-value: RR X P X ; 1 1 g
Bartletts Test Pest Data ## Bartlett's Test g <- length(unique(pest.trt)) N <- length(pest.trt) pest.trt <- factor(pest.trt) sd.trt <- as.vector(tapply(wt.juice, pest.trt, sd)) n.trt <- as.vector(tapply(wt.juice, pest.trt, length)) MS_ERR <- sum((n.trt-1) * sd.trt^2) / (N-g) C.B <- 1 + (1/(3*(g-1))) * (sum(1/(n.trt-1)) - 1/(N-g)) X2.B <- (1/C.B) * ((N-g)*log(MS_ERR) - sum((n.trt-1) * log(sd.trt^2))) p.X2.B <- 1 - pchisq(X2.B, g-1) bart.out <- cbind(X2.B, g-1, qchisq(.95,g-1), p.X2.B) colnames(bart.out) <- c("Bartlett X2", "df", "X2(.95)", "P(>X2)") round(bart.out, 4) > round(bart.out, 4) Bartlett X2 df X2(.95) P(>X2) [1,] 20.8832 10 18.307 0.0219
CRD with Non-Normal Data Kruskal-Wallis Test Extension of Wilcoxon Rank-Sum Test to g > 2 Groups Procedure: Rank the observations across groups from smallest (1) to largest (N = n1+...+ng), adjusting for ties Compute the rank sums for each group: T1,...,Tg . Note that T1+...+Tg = N(N+1)/2
Kruskal-Wallis Test H0: The g population distributions are identical HA: Not all g distributions are identical 12 . .: ( N N 2 T n g = + 3( 1) i T S H N + = 1) 1 i i 2 . .: R R H , 1 g 2 : ( ) P val P H An adjustment to H is suggested when there are many ties in the data. H H t t t = th ' number of observations in group of tied ranks j ( 3 ) j 3 j j j 1 N N
Kruskal-Wallis Test Pest Data ## Kruskal-Wallis Test g <- length(unique(pest.trt)) N <- length(pest.trt) pest.trt <- factor(pest.trt) n.trt <- as.vector(tapply(wt.juice, pest.trt, length)) rank.wj <- rank(wt.juice) ranksum.trt <- as.vector(tapply(rank.wj, pest.trt, sum)) X2.KW <- (12/(N*(N+1))) * sum(ranksum.trt^2/n.trt) - 3*(N+1) p.X2.KW <- 1 - pchisq(X2.KW, g-1) kw.out <- cbind(X2.KW, g-1, qchisq(.95,g-1), p.X2.KW) colnames(kw.out) <- c("Kruskal-Wallis X2", "df", "X2(.95)", "P(>X2)") round(kw.out, 4) > round(kw.out, 4) Kruskal-Wallis X2 df X2(.95) P(>X2) [1,] 46.8797 10 18.307 0
Welchs Test Unequal Variances g n s = = i w w w i i 2 i = 1 i 2 g w y i i g 1 2 = 1 i = (same as last slide) F w y EW i i 1 g w = 1 i ( g ) 1 2 2 2 1 g g 1 w w = = + 1 1 i C m C W W W 2 1 n = 1 i i 1 3 approx ~ = = C F m F F 1, W W W W EW g 2 1 g W
Welchs Test Pest Data WelchF <- function(n, ybar, s2) { g <- length(n) w <- n/s2 sumw <- sum(w) m.W1 <- sum((1/(n-1)) * (1 - w/sumw)^2) m.W <- 1 / (1 + (2*(g-2)/(g^2-1)) * m.W1) nu.W <- 1 / ((3/(g^2-1)) * m.W1) F.EWLS <- (1/(g-1)) * (sum(w*ybar^2) - (sum(w*ybar)^2)/sumw) F.W <- m.W * F.EWLS p.W <- 1 - pf(F.W, g-1, nu.W) Welch.out <- list("F"=F.W, "df1"=g-1, "df2"=nu.W,"P"=p.W) return(Welch.out) } wf.test <- WelchF(n.trt, mean.trt, var.trt) wf.test.out <- cbind(wf.test$F, wf.test$df1, wf.test$df2, wf.test$P) colnames(wf.test.out) <- c("Welch F", "df1", "df2", "P(>F)") round(wf.test.out, 4) Welch F df1 df2 P(>F) 111.5903 10 21.6547 0
Kenward-Roger Method 2 g w y 2 i i g g g 1 1 n s w w 2 = 1 i = = = = 2 1 i i w w w F w y a i i EW i i 2 i 1 1 g w n = = = 1 1 1 i i i i ( ) )( ( 2 ) + + + + 2 2 7 + 2 7 2 + 4 g g + g 21 g = = = c c c ( )( ( ( )( ) ) ) 1 2 3 + + 2 2 2 3 2 5 1 2 3 2 5 1 2 3 2 5 1 g g g g g g g g 1 + 1 2 c a V E a = = = 1 E V 1 KR ( ) ( ) ( ) KR KR 2 2 1 1 g g 1 1 2 c a c a 2 3 KR + 1 g approx ~ = + = = 4 m F m F F KR ( ) ( ) 1, KR KR KR KR EW g 1 1 2 g E KR KR KR
Kenward-Roger Method Pest Data KRF <- function(n, ybar, s2) { g <- length(n) w <- n/s2 sumw <- sum(w) a <- 2 * sum((1/(n-1)) * (1 - w/sumw)^2) c1 <- -21 / (2 * (3*g^2+2*g+5) * (g-1)) c2 <- (7 * (g^2+2)) / (2 * (3*g^2+2*g+5) * (g-1)) c3 <- (7 * (g^2+2*g+4)) / (2 * (3*g^2+2*g+5) * (g-1)) E.KR <- 1 / (1 - a/(g-1)) V.KR <- (2 / (g-1)) * ((1 + c1*a) / ((1-c2*a)^2 * (1-c3*a))) rho <- V.KR / (2 * E.KR^2) nu.KR <- 4 + ((g+1) / ((g-1)*rho - 1)) m.KR <- nu.KR / (E.KR * (nu.KR-2)) F.EWLS <- (1/(g-1)) * (sum(w*ybar^2) - (sum(w*ybar)^2)/sumw) F.KR <- m.KR * F.EWLS p.KR <- 1 - pf(F.KR, g-1, nu.KR) KR.out <- list("F"=F.KR, "df1"=g-1, "df2"=nu.KR,"P"=p.KR) return(KR.out) } krf.test <- KRF(n.trt, mean.trt, var.trt) krf.test.out <- cbind(krf.test$F, krf.test$df1, krf.test$df2, krf.test$P) colnames(krf.test.out) <- c("KR F", "df1", "df2", "P(>F)") round(krf.test.out, 4) KR F df1 df2 P(>F) 104.7307 10 14.087 0
Satterthwaite Method 1 1 1 0 0 1 0 0 ( ) = = L L 1 I 1) Determine a full row rank 1 Contrast matrix ' e.g.: ' g g 1 1 1 g g 1 0 0 1 y 1 1 2 i 1 s n ^ ^ = = = = * C Y Y L L CL L Y 2) Compute: diag ' ' ' F F EW 1 g i y g L ^ ^ ( ) ( 1 ) = Y L CL 3) ' ' 1 with full rank. V g g 1 ^ ^ ^ = = 1 L CL P P 4) Spectral Decomposition: ' ' where eigenvectors and diagonal matrix of eigenvalues of ' ' ' P L CL L CL P P 2 1 1 P L Y g g ( ) ' ' 1 1 1 1 2 = = = = = = * 1 PP P P I Y LP P L Y P L Y P P P ' ' ' ' ' ' ' i F 1 1 i g 1 1i 1 g g g = = 1 1 i i i ^ ^ = = = = th P L Y P L CLP P P P P 5) ' ' ' ' ' ' where ' ' ' where is the column of V i P P P P P e e I 1 1 1 i i i i i i i g i i g P L Y ' ' ^ = = P L Y e e ' ' ' ~ Recall that spectral decomposition taken on the Estimated variance-covariance matrix i V t i i i i i i ^ ^ ^ ^ 2 2 i 2 i 2 2 ^ i i i i 2 Consider: ~ 1 2 i i i i E V V i i i i i 2 i ^ i V i i i i i
Satterthwaite Method ( ) ( ) ( ( ) ) 2 i 2 i 4 i 1 1 2 1 1 n S n S n 4 i 2 n S V S V S ^ ( ) = = = i i i 2 n 2 i 2 i 6) ~ 2 1 V n 1 i 2 2 i 2 i 1 n i i i 2 ^ P L CLP ' ' g i i V S ^ ^ ^ ^ ^ ^ ^ = 2 j P L CLP P L CLP approx approx ' ' which by delta method: approx ' ' V V V i i i i i 2 j S = 1 j 2 ^ C ^ C 4 j S 2 i g 1 n S n = = = P L LP e e 2 ' ' where diag ' i i j j 2 j 2 j 2 j 1 S n S S = 1 j j i j 2 2 ^ C ( ) 2 4 j 4 j 4 j P L e S S S ' ' n g g g 1 n = = i i P L LP P L e 2 ' ' 2 ' ' 2 i i i i 2 j 2 j 1 1 1 S n n n = = = 1 1 1 j j j j j j j 2 ^ 2 i 2 2 i 7) ( ) i 2 ^ 4 j P L e S ' ' n g V i i 2 i 2 j 1 n = 1 j j 2 2 P L Y P L Y 1 g ' ' ' ' 1 E t E F = = * * 2 8) ~ 2 i S i i F F E 1, g S 1 2 2 g S i = 1 i S i i i 1 g 2 i 1 g 2 1 E F = 1 i * where if any 2, they are replaced with 0 in computation i S i S i 1 g 1 2 2 g ( ) = 1 i 1 g i i S 2 = 1 i i
Satterthwaite Method Pest Data SattF <- function(n, ybar, s2, Lp) { g <- length(n) C <- diag(s2/n) LCL <- Lp %*% C %*% t(Lp) lambda <- eigen(LCL)$values P <- eigen(LCL)$vectors Fstar <- (1/(g-1)) * t(ybar) %*% t(Lp) %*% P %*%solve(diag(lambda)) %*% t(P) %*% Lp %*% ybar delta1 <- lambda^2 PLe <- t(P) %*% Lp s4_n2nm1 <- matrix(s2^2 / (n^2 * (n-1)), ncol=1) var.lambda <- 2 * ((PLe^4) %*% s4_n2nm1) delta <- 2 * lambda^2 / var.lambda sum.delta_deltam2 <- 0 for (i in 1:(g-1)) { if (delta[i] > 2) sum.delta_deltam2 <- sum.delta_deltam2 + delta[i] / (delta[i] - 2) } nu.S <- (2 * sum.delta_deltam2) / (sum.delta_deltam2 - (g-1)) if (nu.S <= 0) nu.s <- 1 if (nu.S > sum(n) - g) nu.S <- sum(n) - g p.S <- 1 - pf(Fstar, g-1, nu.S) Satt.out <- list("F"=Fstar, "df1"=g-1, "df2"=nu.S,"P"=p.S) return(Satt.out) }
Satterthwaite Method Pest Data L <- matrix(cbind(rep(-1,length(mean.trt)-1), diag(length(mean.trt)-1)), ncol=length(mean.trt)) sattf.test <- SattF(n.trt, mean.trt, var.trt, L) sattf.test.out <- cbind(sattf.test$F, sattf.test$df1, sattf.test$df2, sattf.test$P) colnames(sattf.test.out) <- c("Satt F", "df1", "df2", "P(>F)") round(sattf.test.out, 4) Satt F df1 df2 P(>F) 142.5093 10 9.5002 0