Microbial Redox Reactions in Sulfate Reduction and Methanogenesis
Explore the decoupling of redox reactions between carbonate and acetate, carbonate and methane, and sulfate and sulfide to support sulfate reducers and methanogens with energy. Learn how to specify starting fluid composition, add reactants, identify and define microbes, set reaction intervals, define inlet fluid properties, specify domain size and flow rate, and set mass transport properties in a comprehensive model.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
Config Redox Couples Decouple reaction between carbonate and acetate, carbonate and methane, and sulfate and sulfide. Decoupling provides for the redox disequilibrium that supplies sulfate reducers and methanogens with energy. Default setting: all redox reactions are coupled.
Specify domains starting fluid composition on the Initial pane After decoupling redox pairs, you can add carbon and sulfur species in multiple redox states. Ca-HCO3 water containing SO42 , pH = 6
You add reactants and enable redox kinetics on the Reactants pane. Click to expand Rate of acetate addition to the aquifer SO4 reducers Methanogens add Simple Aqueous CH3COO add Simple Aqueous Ca++ add Kinetic Microbial reaction (2X)
Identify each microbe with a label and reaction. Microbes can compete for common substrates or carry out separate reactions. Specify kinetic, thermodynamic, and growth parameters for each microbe. Initial biomass Growth yield and decay constant Rate constant, half- saturation constants ATP energy, ATP number Form of Monod terms for e- donating (D) and accepting (A) half-reactions
Define the reaction intervals. Specify what fluids flow into the domain, and when. A single inlet fluid flows into the domain for 105 years.
The inlet fluid is defined on the Fluids pane The inlet fluid is identical to the Initial fluid
Specify domain size and gridding on the Domain pane. Domain is 200 km long, divided into 50 nodal blocks
Specify flowrate on the Flow pane. Set specific discharge or head drop
Set various mass transport properties on the Medium pane. Run Go traces the model
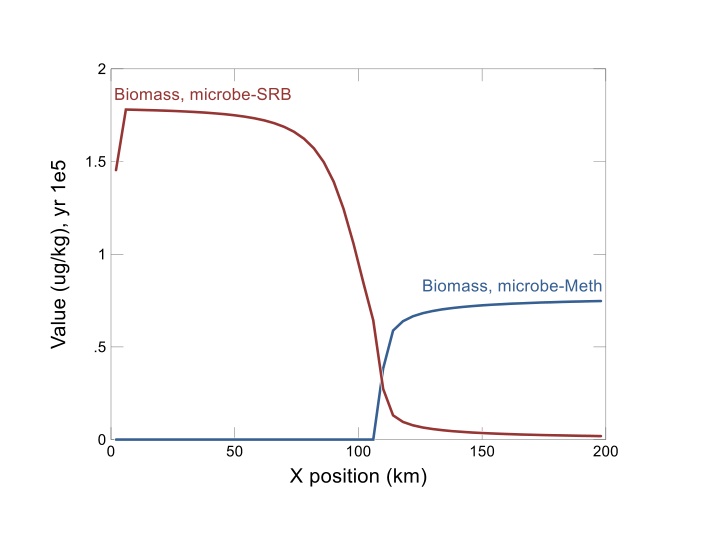
After 105 years of metabolism, growth, and decay, two microbiological zones develop in the aquifer.
SRB dominate where sulfate is abundant. Methane accumulates in second zone of the aquifer.