PCR - History, Introduction, Basic Constituents, and Procedure
PCR, invented by Kary Mullis, is a vital nucleic acid amplification method that utilizes DNA polymerase to copy strands of DNA. The process involves amplifying selected DNA sequences with synthetic primers and key constituents like Taq polymerase and dNTPs. Thermal cycling in a PCR machine involves denaturation, primer annealing, and DNA extension steps to achieve replication. Explore the origins, significance, and essential components of PCR in this comprehensive guide.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
PCR (Polymerase Chain Reaction) Dr. Abdul Haque 1
History PCR was invented by Kary Banks Mullis (December 28, 1944 August 7, 2019) was an American biochemist. He received the 1993 Nobel Prize in Chemistry. Dr. Abdul Haque 2
In 1983, Mullis was working for Cetus Corporation as a chemist. While driving in the vicinity of his country home in Mendocino County he had the idea of the mechanism of PCR. He was supported by his seniors but there was no practical success because polymerases he used were not heat stable. In 1986, one of his colleagues, Randall Saiki used Thermophilus aquaticus (Taq) DNA polymerase to solve this problem. Dr. Abdul Haque 3
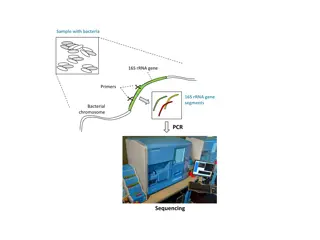
INTRODUCTION PCR is the best-developed and most widely used method of nucleic acid amplification. -- It is based on the ability of DNA polymerase to copy a strand of DNA by elongation of complementary strands initiated by a pair of chemically synthesized oligonucleotide primers. -- The basic technique of PCR includes repeated cycles of amplifying the selected nucleic acid sequences. -- Dr. Abdul Haque 4
Some parts of the sequence of a DNA molecule are unique. -- These are termed as signature sequences . In PCR, these signature sequences are targeted for amplification. -- In diagnostic PCR, detection of these sequences means detection of DNA of interest. --
Basic Constituents of a PCR reaction Primers These are a pair of synthetic oligonucleotides which anneal to sites flanking the region to be amplified (signature sequences). Polymerase An enzyme that extends the primers to form a new strand. Taq polymerase is most commonly used.
dNTPs A mixture of dGTP (dATP, dTTP, and dCTP). It is the construction material used for a new strand. Mg++ Cements the bases together. K+ Provides an environment (pH) conducive for PCR reaction.
Procedure in Thermal Cycler Each cycle consists of three steps: 1. DNA denaturation : The double strands of the target DNA are separated at 95 C. Time varies according to the size of amplification product. 2. Primer annealing : It is performed at a lower variable temperature (according to GC content of DNA), in which primers anneal to their complementary target sequences. Time varies according to the size of amplification product. Dr. Abdul Haque 8
3. Extension: DNA polymerase extends the sequences starting from the primers. The temperature depends on the Polymerase used. For Taq polymerase, the temperature is 72 C. Dr. Abdul Haque 9
At the end of each cycle (each consisting of the above three steps), the quantities of PCR products are theoretically doubled. -- Generally, performance of 20-30 thermal cycles results in an exponential increase (nearly 1 million fold) in the total number of DNA copies synthesized. -- The amplified products are traditionally detected by agarose gel electrophoresis followed by staining with ethidium bromide. -- Dr. Abdul Haque 11
How the amplification product size is defined? -- There are two primers, one each for one single strand of the double stranded DNA to be amplified. -- During denaturation (first step), the two strands are separated and each primer attaches to its respective strand during annealing (2nd step). -- During extension (3rd step), the polymerase start building new strands from the point where a primer has attached. Dr. Abdul Haque 14
So these new strands have one defined end (the starting point) but their other end is undefined so the length is variable. -- -- During the next cycle when new strands will be formed, there will be complementary strands of those strands whose one end is already defined. Now the other primer will attach to these complementary strands. -- Dr. Abdul Haque 15
-- In this way second end of the product will also be defined, and when extension will occur, the right size of product will be obtained. -- As a result, after 4-5 cycles, the desired size of amplified product is obtained. Dr. Abdul Haque 16
Why we cannot perform PCR on RNA? There are two reasons: Usually used polymerases do not recognize uracil (however there are some polymerases from archeae which an recognize uracil but they are very expensive). -- RNA is generally unstable at higher temperatures. -- Dr. Abdul Haque 17
PCR Controls Inclusion of control reactions is essential for monitoring the success of PCR reactions. Positive control: Wherever possible, a positive control should be included to check that the PCR conditions used can successfully amplify the target sequence. It is a known DNA which is routinely amplified by using these PCR conditions. Dr. Abdul Haque 18
Negative control PCR is extremely sensitive, so a negative control containing no template DNA should always be included to check for contamination. Dr. Abdul Haque 19
Laboratory conditions PCR setup should be performed in a separate area from PCR analysis to ensure that reagents used for PCR do not become contaminated with PCR products. Similarly, pipets used for analysis of PCR products should never be used for setting up PCR. Dr. Abdul Haque 20
TYPES OF PCR Dr. Abdul Haque 21
REVERSE TRANSCRIPTASE (RT)-PCR RT-PCR was developed to amplify RNA targets. -- In this process, RNA targets are first converted to complementary DNA (cDNA) by RT, and then amplified by PCR. -- RT-PCR has played an important role in diagnosing RNA- containing virus infections such as HCV, Dengue virus. -- Dr. Abdul Haque 22
NESTED PCR -- Nested PCR, designed mainly to increase sensitivity (detect smaller quantities of target), uses two sets of amplification primers. -- One set of primers is used for the first round of amplification, which consists of 15 to 30 cycles. The amplicons are then subjected to a second round of amplification with new set of primers specific for an internal sequence that was amplified by the first primer pair. -- Dr. Abdul Haque 23
Nested PCR has extremely high sensitivity because of the dual amplification process. -- It also has high specificity because the second primer set verifies the specificity of the first-round product. -- Dr. Abdul Haque 25
MULTIPLEX PCR Multiplex PCR is an amplification reaction in which two or more sets of primer pairs specific for different targets are introduced in the same tube. -- -- Thus, more than one unique target DNA sequence in a specimen can be amplified at the same time. -- This co-amplification of multiple targets can be used to detect multiple pathogens from a single specimen. Dr. Abdul Haque 26
RAPD: Rapid amplified polymorphic DNA analysis RAPD is a PCR-based tool enabling the study of organisms at the molecular level. It uses small, nonspecific primers to amplify seemingly random regions of genomic DNA. Successful primer pairs produce different banding profiles of PCR products between individuals, strains, species, etc., when analyzed using an agarose gel. In RAPD, the primers are only ~10 bases long. As a result, annealing temperatures required are <40 C. Dr. Abdul Haque 28
Long-range PCR PCR products of up to 4 kb can be routinely amplified using standard PCR protocols using Taq DNA polymerase. However, amplification of PCR products longer than 4 kb often fails without lengthy optimization. Reasons for failure include nonspecific primer annealing, secondary structures in the DNA template, and suboptimal cycling conditions. Dr. Abdul Haque 30
But the main reason is DNA damage (such as DNA depurination, i.e., release of adenine or guanine) as a single DNA lesion within the template will stop PCR. DNA damage during PCR cycling can be minimized with specific buffering substances that stabilize the pH of the reaction. Commercial kits are usually used. Dr. Abdul Haque 31
Single-cell PCR Single-cell PCR provides a valuable tool for genetic characterization using a limited amount of starting material. Individual cells of interest can be isolated based on cell- surface markers or physical appearance by flow cytometry or micromanipulation. A PCR system that is highly efficient, specific, and sensitive is required. Commercial kits are available. Dr. Abdul Haque 32
In situ PCR In situ PCR is a PCR reaction that occurs inside the cell on a slide, thus combining the sensitivity of PCR or RT-PCR amplification with in situ hybridization. In situ PCR allows cellular markers to be identified and further enables the localization to cell-specific sequences within cell populations, such as tissues and blood samples. Therefore, it is a powerful tool in applications such as the study of disease progression. Dr. Abdul Haque 33
Fresh or fixed cells or tissue samples can be used in the procedure. Preparation of the sample is critical to the result, with fixation having a direct influence on PCR signal. The procedure is suitable for use with radiolabeled, fluorescently labeled or biotin-labeled nucleic acid probes. The PCR process is essentially the same as a standard PCR, but with some modified reaction conditions (e.g., Mg2+ concentration). Dr. Abdul Haque 34
Hot start PCR Hot Start PCR is a technique that reduces non-specific amplification. In Hot Start PCR, the polymerase (or dNTPs or Mg2+) are added when the mixture has reached a temperature higher than Ta (annealing temperature) and then PCR cycles are started. Dr. Abdul Haque 35
High-fidelity PCR High-fidelity PCR, utilizes a DNA polymerase with a low error rate (e.g. Pfu from Pyrococcus furiosus) and results in a high degree of accuracy in the replication of the DNA of interest. Dr. Abdul Haque 36
Touch down PCR In touchdown PCR the temperature selected for the annealing step is initially set 5 C-10 C higher than the calculated Tm (melting temperature) of the primers. Then progressively transitions to a lower, more permissive annealing temperature over the course of successive cycles. Tm is the temperature at which one half of the DNA duplex will dissociate to become single stranded. Dr. Abdul Haque 37
REAL-TIME PCR This method is based on using fluorescent labeled probes to detect, confirm, and quantify the PCR products as they are being generated in real time. SYBR-Green labelled probes Real-time fluorescence is monitored by using fluorescent dyes such as SYBR-Green, which binds non-specifically to double-stranded DNA generated during the PCR amplification. -- Dr. Abdul Haque 38
SYBR Green labeling Dr. Abdul Haque 39
TaqMan fluorescent probes -- TaqMan fluorescent probes bind specifically to amplification target sequences but are more expensive. The probes are labelled by a reporter dye alongside a quencher molecule. -- -- The purpose of quencher molecule is to keep the fluorescence at a very low level. When polymerase reaches the bound probe it removes it and in the process, the probe is broken. -- When reporter dye moves away from quencher molecule, it generates a high level signal which is recorded. -- Dr. Abdul Haque 40
Molecular Beacons Molecular beacons have a characteristic stem-loop structure through which the 5 and 3 ends are maintained in close proximity. -- -- Fluorescence from a fluorochrome at one end of the probe is suppressed by a nearby Quencher. However, unlike TaqMan probes molecular beacons are not degraded during the amplification process. -- They bind to the target in every cycle in order to produce measurable fluorescence. -- Dr. Abdul Haque 42
The real-time PCR systems not only reduce the detection time (results can be ready in less than on hour), but also can reduce contamination risks because amplification and detection occur within a closed system. -- Dr. Abdul Haque 46
Applications of PCR 1. The diagnosis of infectious agents, including HIV, hepatitis, malaria, anthrax, etc. 2. Information on a patient s prognosis by detecting small mutations in certain genes of cancerous cells. 3. The analysis of mutations that occur in many genetic diseases (e.g. cystic fibrosis). 4. In forensics laboratories to target a tiny amount of original DNA, e.g., from a droplet of blood or a single hair. Dr. Abdul Haque 47
5.To generate large amounts of pure DNA from tiny amount of template strand during cloning. 6.To explore relationships among species in the field of evolutionary biology. 7.To understand the ancient human migration patterns in anthropology. 8.To spot the ancient human race in archaeology. 9.To amplify DNA from extinct species or cryopreserved fossils of millions of years. Dr. Abdul Haque 48