Polymerase Chain Reaction: Amplifying DNA Samples Efficiently
Polymerase Chain Reaction (PCR) is a powerful technique used to amplify small samples of DNA, enabling the production of large amounts for analysis. By repeatedly cycling through steps of DNA separation, primer binding, and replication, PCR can create multiple copies of a target DNA sequence. This process is crucial in various fields such as genetics, forensics, and biomedical research. Learn more about the PCR process, its applications, and the importance of primer design in this comprehensive overview.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
02 March 2025 Polymerase Chain Reaction Aims To understand the process of PCR and its uses. Starter - Match each term with its correct description (work in pairs)
Animations http://www.dnalc.org/ddnalc/resource s/pcr.html http://www.maxanim.com/genetics/PC R/pcr.swf
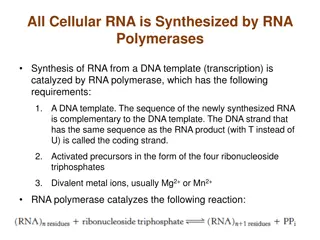
Information Polymerase chain reaction enables large amounts of DNA to be produced from very small samples (0.1ml) There is a repeating cycle of: separation of double DNA strands synthesis of a complementary strand for each
What happens? Sample DNA , nucleotides, DNA primers & thermostable DNA polymerase placed in PCR machine. Strands of sample DNA separated by heating to 95oC Mixture cooled to 37oC to allow primers to bind. Mixture heated to 72oC for replication (optimum temp of DNA polymerase) Cycle repeats many times (~8mins /cycle)
Problems Separation achieved by heating to 95oC no suitable helicase DNA polymerase can t work on completely single stranded DNA double stranded regions needed at the start of sequence to be copied: primers (short sequences DNA) complementary to bases at start of region to be copied used To synthesize primers , base sequence at start must be known
TTAACGGGGCCCTTTAAA.....TTTAAACCCGGGTTT AATTGCCCCGGGAAATTT..... AAATTTGGGCCCAAA the sequence of bases which ONLY flank a particular region of a particular organism's DNA, and NO OTHER ORGANISM'S DNA. This region would be a target sequence for PCR.
The first step for PCR would be to synthesize "primers" of about 20 letters- long ONE primer exactly like the lower left- hand sequence, and ONE primer exactly like the upper right-hand sequence: TTAACGGGGCCCTTTAAA..... TTTAAACCCGGGTT AATTGCCCCGGGAAATTT.......................> and: <........................................TTTAAACCCGGGTTT AATTGCCCCGGGAAATTT........AAATTTGGGCCCAAA
DNA polymerase must be thermostable (37oC 95oC used) to avoid fresh enzyme being added.
Remember. Nucleotides used must be very pure. DNA must not be contaminated - any foreign DNA would also be copied .PCR in legal cases in UK suspended at present
Uses of PCR http://nobelprize.org/nobel_prizes/che mistry/laureates/1993/illpres/already .html
This powerpoint was kindly donated to www.worldofteaching.com http://www.worldofteaching.com is home to over a thousand powerpoints submitted by teachers. This is a completely free site and requires no registration. Please visit and I hope it will help in your teaching.