Protein Structure Prediction and Bioinformatics
Learn about protein structure prediction methods such as homology modeling, bioinformatics tools, and using physics in predicting protein folding. Explore the interplay between amino acid sequences and protein structures.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
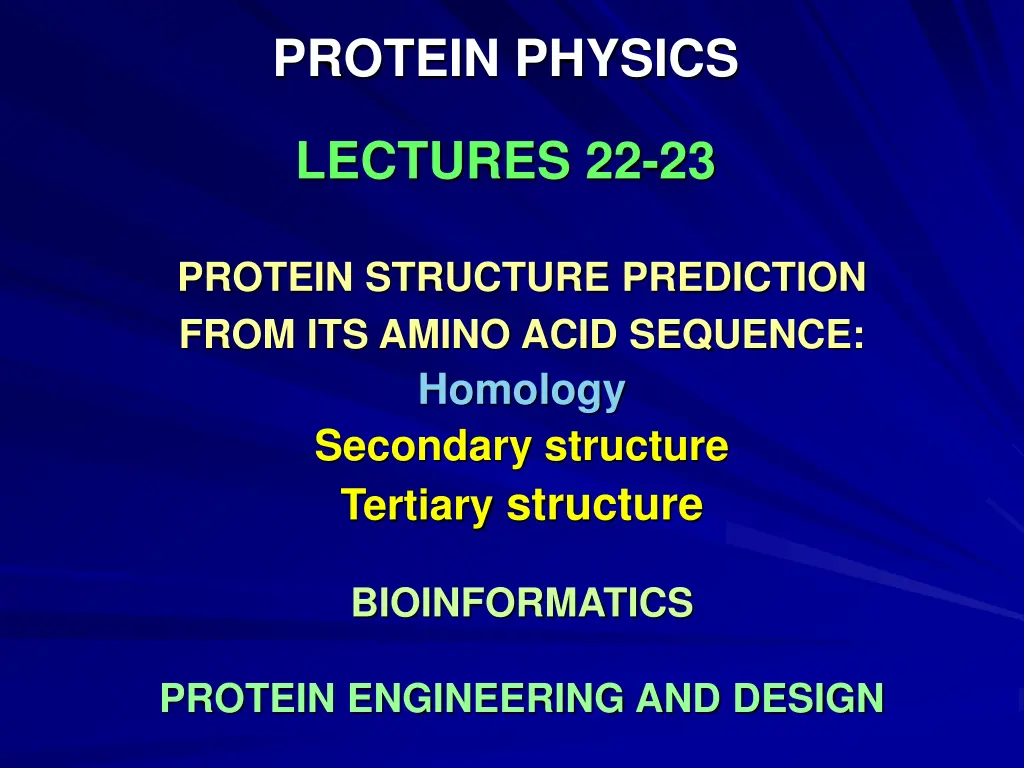
PROTEIN PHYSICS LECTURES 22-23 PROTEIN STRUCTURE PREDICTION FROM ITS AMINO ACID SEQUENCE: Homology Secondary structure Tertiary structure BIOINFORMATICS PROTEIN ENGINEERING AND DESIGN
Homology -- -- --
PREDICTION FROM HOMOLOGY SIMILAR SEQUENCES SIMILAR FOLDS ______ __________________ _______ ACCURACY = t/(t+f) SEQUENCE ALIGNMENT: SMITH-WATERMAN, BLAST,
N0 ======= GOOD PREDICTION ======= TWILIGHT
PREDICTION FROM HOMOLOGY cytochromes c High homology: TWILIGHT ZONE : 10-25% IDENTITY: Low but existing homology. Sequence identity: 15% NO homology. Sequence identity: 15%
Multiple homology key sites (core, 20 structure, active site) PROFILES HIDDEN MARKOV MODELS (HHMer) etc. BIOINFORMATICS
Multiple homology PROFILE TARGET...APGDEFG--HIKKLMAATC... SEQUENCE
PREDICTION FROM PHYSICS: PROTEIN CHAIN FOLDS SPONTANEOUSLY SEQUENCE HAS ALL INFO TO PREDICT: 2O STRUCTURE, 3D STRUCTURE, SIDE CHAIN ROTAMERS, S-S BONDS, etc.
PREDICTION FROM PHYSICS (OR PROTEIN STATISTICS) 2O STRUCTURES
no (Gly): coil , 1 : , , coil -np- --p-- Pro , 2 : (3 rot.) imino: coil, turn, N
PREDICTION FROM PROTEIN STATISTICS (OR PHYSICS) TEMPLATE OF SUPER-SECONDARY STRUCTURE np Gly -
FLUCTUATING SECONDARY STRUCTURE IN UNFOLDED POLYPEPTIDE CHAIN OLYGO- & POLYPEPTIDES ALB
FLUCTUATING SECONDARY STRUCTURE AT THE AVERAGE SURFACE OF A GLOBULE PHD, JPRED, PSIPRED, bpti ALB
Prediction, 1985 X-ray str.,1990 A B C D .---different---
THREADING BIOINFORMATICS
THREADING: CORRECT FOLD, BAD ALIGNMENT
ENGINEERING & DESIGN DeGrado, 1989 DOES NOT MELT ! MOLTEN GLOBULE + IONBINDING SOLID
Ptitsyn Dolgikh Finkelstein Fedorov Kirpichnikov 1987-97 Albebetin, Albeferon,
non-polar: core polar: surface
