Understanding Randomized Complete Block Design (RCBD)
Learn about Randomized Complete Block Design (RCBD), a statistical design used in experiments to reduce variation among experimental units. Discover how blocks, fixed and random effects, missing value imputation, and power analysis play crucial roles in RCBD experiments.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
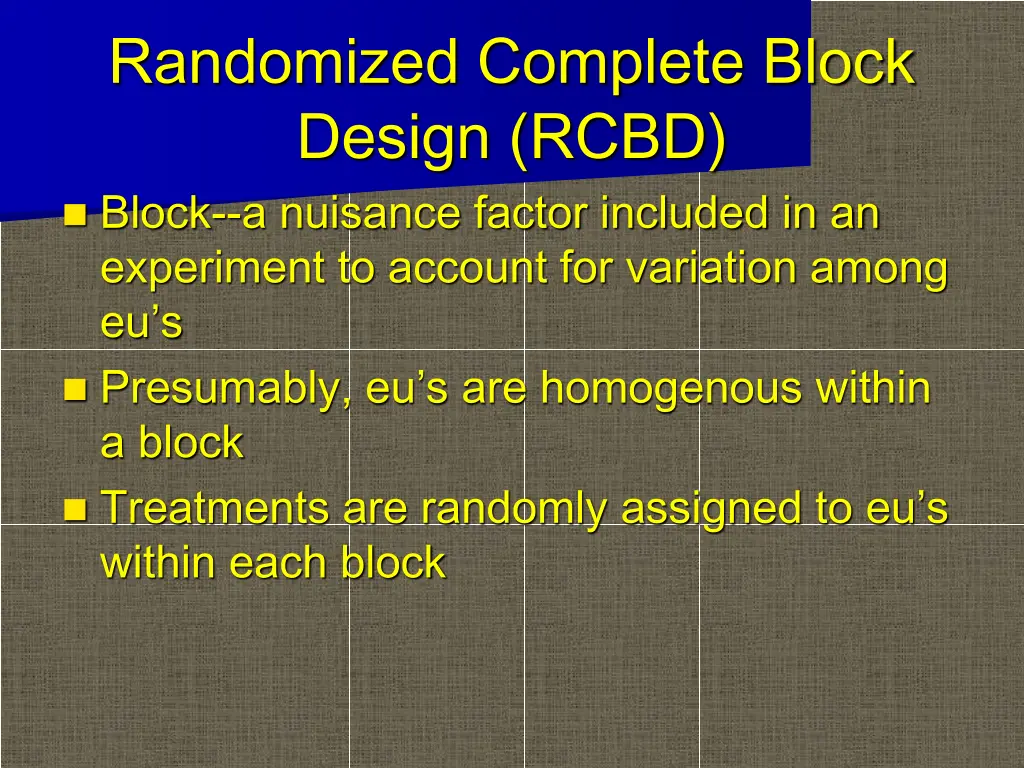
Randomized Complete Block Design (RCBD) Block--a nuisance factor included in an experiment to account for variation among eu s Presumably, eu s are homogenous within a block Treatments are randomly assigned to eu s within each block
RCBD The model and hypotheses , 0 ( = + + + N + , Y ij i j ij ij + 2 ) iid ij ij : 0 H o i
Blocks in RCBDs Blocks can be modeled as both fixed and random effects (Soil example) Block: Soil type (fixed or random?) Treatment: Nitrogen x Watering Regimen Response: IR/R reflection
RCBD Discussion There is some controversy as to whether fixed block effects should be tested F test is considered at best approximate Additivity of the block and factor effects Error includes lack-of-fit Practical considerations Both block and factor could have a factorial structure
Missing values in RCBDs Missing values result in a loss of orthogonality (generally) A single missing value can be imputed The missing cell (yi*j*=x) can be estimated by profile least squares ' . * a + ' b ' ay by y . * .. i j = x ( )( ) 1 1
Imputation The error df should be reduced by one, since x was estimated SAS can compute the F statistic, but the p- value will have to be computed separately The method is efficient only when a couple cells are missing
Alternate Imputation approaches The usual Type III analysis is available, but be careful of interpretation Little and Rubin use MLE and simulation- based approaches PROC MI in SAS v9 implements Little and Rubin approaches
Power analysis Power calculations change little b replaces n in formulas 2 bL = = For H : , 0 use L o 2 2 ic 2 i b , 0 = = For H : use o 2 The error df is (a-1)(b-1)