Validating MHC Folding on Yeast: Constructs Representation and Library Creation
Plasmid and surface representations of MHCI and MHCII constructs for yeast display are illustrated, along with a flow chart for validation. The process involves identifying MHC and TCR, testing for binding, and creating error-prone libraries. Flow cytometry plots and selection data demonstrate successful enrichment processes. Amplicon sequencing formatting is also showcased for cluster identification.
Uploaded on Mar 04, 2025 | 1 Views
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
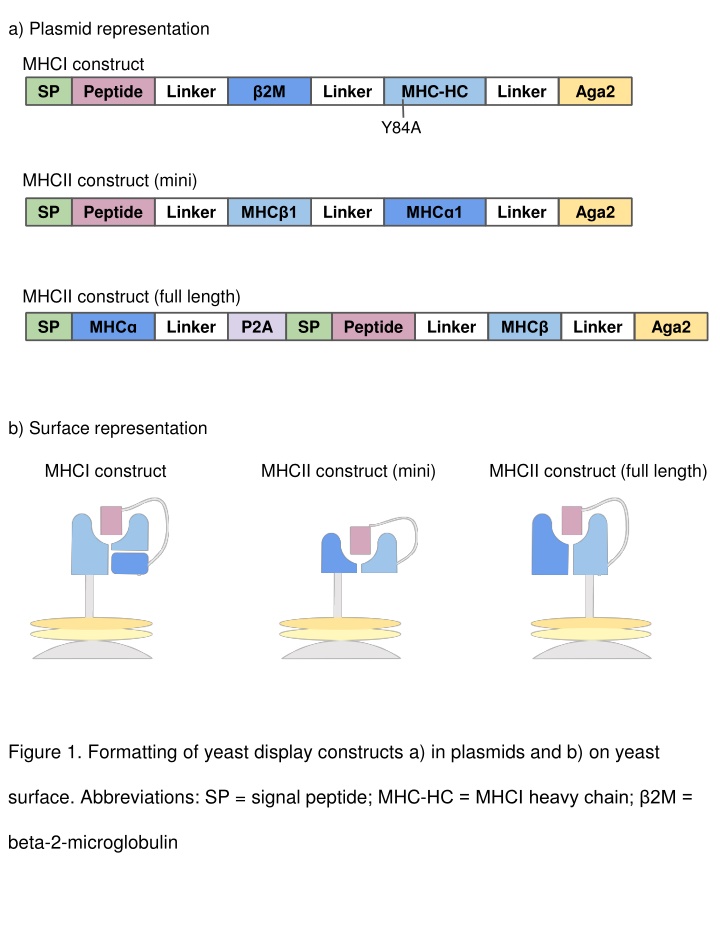
a) Plasmid representation MHCI construct Peptide SP Linker Linker MHC-HC Linker Aga2 2M Y84A MHCII construct (mini) SP Peptide Linker Linker Linker Aga2 MHC 1 MHC 1 MHCII construct (full length) SP Linker P2A SP Peptide Linker Linker Aga2 MHC MHC b) Surface representation MHCI construct MHCII construct (mini) MHCII construct (full length) Figure 1. Formatting of yeast display constructs a) in plasmids and b) on yeast surface. Abbreviations: SP = signal peptide; MHC-HC = MHCI heavy chain; 2M = beta-2-microglobulin
Validating MHC Folding on Yeast Identify MHC and TCR of interest. Has this MHC previously been expressed on yeast? Yes No Proceed with making library. Does this MHC have a known TCR+peptide ligand? Yes No Express pMHC on yeast and test for binding (tetramer staining or tag enrichment). Is there binding? Cannot determine if MHC is folded. Yes No Make error prone library and select. Install stabilizing mutations. Proceed with making library. Figure 2. Flow chart for validating a yeast display construct.
Figure 3. Example flow cytometry plots showing Myc tag staining pre- and post- selection with 400nM TCR-loaded streptavidin beads. The left column shows a successfully enriched population of pMHC-expressing yeast, suggesting that the construct is properly folded on the surface of the yeast. The right column shows an unsuccessful enrichment, suggesting that the construct is not properly folded.
a) b) Figure 4. Sample selection data for TCR deorphanization. a) Myc epitope tag staining shows enrichment over successive rounds of selection (gated on unstained yeast). b) Selection scheme and data for five rounds of selection on a randomized 9mer library displayed by mouse MHCI H-2Db. In this example, spot check of a dozen colonies following round 3 indicated enrichment of three peptides.
i5 Anchor F Primer R Primer BC i7 Anchor Peptide NNNNNN NNNNNN Figure 5. Amplicon sequencing formatting. Short randomized sequences for cluster identification are indicated as NNNNNN . Abbreviations: F primer = Forward NGS primer; R primer = Reverse NGS primer; BC = Index Barcode; N indicates any nucleotide